[English] 日本語
 Yorodumi
Yorodumi- PDB-7r1l: Clostridium thermocellum CtCBM50 structure in complex with beta-1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7r1l | ||||||
|---|---|---|---|---|---|---|---|
| Title | Clostridium thermocellum CtCBM50 structure in complex with beta-1,4-GlcNAc trisaccharide | ||||||
 Components Components | Spore coat assembly protein SafA | ||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / CBM50 / LysM / carbohydrate-binding / GlcNAc | ||||||
| Function / homology |  Function and homology information Function and homology informationSpore coat assembly protein SafA / CAP domain, YkwD-like / CAP domain / Cysteine-rich secretory protein family / CAP superfamily / Lysin motif / LysM domain superfamily / LysM domain / LysM domain profile. / LysM domain Similarity search - Domain/homology | ||||||
| Biological species |  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.45 Å MOLECULAR REPLACEMENT / Resolution: 1.45 Å | ||||||
 Authors Authors | Ribeiro, D.O. / Costa, R. / Palma, A.S. / Carvalho, A.L. | ||||||
| Funding support |  Portugal, 1items Portugal, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Unravelling Clostridium thermocellum LysM domains: a molecular view on the cooperative recognition of chitin and peptidoglycan Authors: Ribeiro, D.O. / Palma, A.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7r1l.cif.gz 7r1l.cif.gz | 46.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7r1l.ent.gz pdb7r1l.ent.gz | 31.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7r1l.json.gz 7r1l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7r1l_validation.pdf.gz 7r1l_validation.pdf.gz | 820.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7r1l_full_validation.pdf.gz 7r1l_full_validation.pdf.gz | 823.2 KB | Display | |
| Data in XML |  7r1l_validation.xml.gz 7r1l_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  7r1l_validation.cif.gz 7r1l_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r1/7r1l https://data.pdbj.org/pub/pdb/validation_reports/r1/7r1l ftp://data.pdbj.org/pub/pdb/validation_reports/r1/7r1l ftp://data.pdbj.org/pub/pdb/validation_reports/r1/7r1l | HTTPS FTP |
-Related structure data
| Related structure data |  5k2lS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 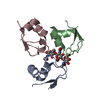
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 6474.537 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Acetivibrio thermocellus (bacteria) / Gene: Cthe_0300 / Production host: Acetivibrio thermocellus (bacteria) / Gene: Cthe_0300 / Production host:  #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | #3: Chemical | ChemComp-ACT / | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.28 Å3/Da / Density % sol: 46.1 % |
|---|---|
| Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, sitting drop / pH: 4.6 Details: 0.1 M sodium acetate buffer pH 4.6, 2 M ammonium sulphate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID29 / Wavelength: 0.9677 Å / Beamline: ID29 / Wavelength: 0.9677 Å |
| Detector | Type: PSI JUNGFRAU 4M / Detector: PIXEL / Date: Jul 9, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9677 Å / Relative weight: 1 |
| Reflection | Resolution: 1.45→49.34 Å / Num. obs: 30139 / % possible obs: 97.1 % / Redundancy: 8.6 % / CC1/2: 1 / Rmerge(I) obs: 0.048 / Rpim(I) all: 0.025 / Net I/σ(I): 20.9 |
| Reflection shell | Resolution: 1.45→1.48 Å / Redundancy: 6.8 % / Rmerge(I) obs: 0.735 / Num. unique obs: 1427 / CC1/2: 0.81 / Rpim(I) all: 0.468 / % possible all: 91.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5K2L Resolution: 1.45→49.34 Å / Cor.coef. Fo:Fc: 0.968 / Cor.coef. Fo:Fc free: 0.959 / SU B: 1.035 / SU ML: 0.04 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.062 / ESU R Free: 0.064 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 85.44 Å2 / Biso mean: 23.344 Å2 / Biso min: 12.4 Å2
| ||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.45→49.34 Å
| ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.452→1.49 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller


 PDBj
PDBj






