+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7qjf | ||||||
|---|---|---|---|---|---|---|---|
| Title | Llp mutant C1G, lytic conversion lipoprotein of phage T5 | ||||||
 Components Components | Lytic conversion lipoprotein | ||||||
 Keywords Keywords | VIRAL PROTEIN / Phage protein / Periplasmic protein / Soluble of acylated WT protein STRUCTURE FROM UNIO / UNIO VERSION 2.9.5 | ||||||
| Function / homology | negative regulation of receptor-mediated virion attachment to host cell / superinfection exclusion / host cell plasma membrane / membrane / Lytic conversion lipoprotein Function and homology information Function and homology information | ||||||
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Degroux, S. / Mestdach, E. / Vives, C. / Le Roy, A. / Salmon, L. / Herrman, T. / Breyton, C. | ||||||
| Funding support |  France, 1items France, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Llp mutant C1G, lytic conversion lipoprotein of phage T5 Authors: Degroux, S. / Mestdach, E. / Vives, C. / Le Roy, A. / Salmon, L. / Herrman, T. / Breyton, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7qjf.cif.gz 7qjf.cif.gz | 377 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7qjf.ent.gz pdb7qjf.ent.gz | 312.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7qjf.json.gz 7qjf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7qjf_validation.pdf.gz 7qjf_validation.pdf.gz | 399.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7qjf_full_validation.pdf.gz 7qjf_full_validation.pdf.gz | 520.7 KB | Display | |
| Data in XML |  7qjf_validation.xml.gz 7qjf_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  7qjf_validation.cif.gz 7qjf_validation.cif.gz | 37.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qj/7qjf https://data.pdbj.org/pub/pdb/validation_reports/qj/7qjf ftp://data.pdbj.org/pub/pdb/validation_reports/qj/7qjf ftp://data.pdbj.org/pub/pdb/validation_reports/qj/7qjf | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 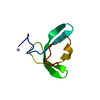
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7052.991 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Mutated version of the lytic conversion lipoprotein produced from a MBP-Llp fusion protein with TEV cleavage site. The mature protein results in the same protein sequence as the WT protein, ...Details: Mutated version of the lytic conversion lipoprotein produced from a MBP-Llp fusion protein with TEV cleavage site. The mature protein results in the same protein sequence as the WT protein, with a Glycine in amino acide +1 in place of the acylated cysteine. Source: (gene. exp.)  Escherichia phage T5 (virus) / Gene: llp, T5.158, T5p154 / Plasmid: pMAL-p2E / Production host: Escherichia phage T5 (virus) / Gene: llp, T5.158, T5p154 / Plasmid: pMAL-p2E / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 25 mM no Tris, 75 mM no NaCl, 1.024 mM 13C 15N Sol-Llp, 90% H2O/10% D2O Details: Sol-Llp protein is soluble and the buffer is 25 mM Tris pH6.5, 75 mM NaCL. Label: 15N_C13_Sol-Llp / Solvent system: 90% H2O/10% D2O | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||
| Sample conditions | Ionic strength: 75 mM / Label: 15N_C13_Sol-Llp / pH: 6.5 / Pressure: 1 atm / Temperature: 303 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 4 | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 80 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller



 PDBj
PDBj HNCA
HNCA