[English] 日本語
 Yorodumi
Yorodumi- PDB-7q3v: Re-refined structure of a type III antifreeze protein isoform HPLC 12 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7q3v | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Re-refined structure of a type III antifreeze protein isoform HPLC 12 | ||||||||||||
 Components Components | Type-3 ice-structuring protein HPLC 12 | ||||||||||||
 Keywords Keywords | ANTIFREEZE PROTEIN / restrained MD refinement scheme / Amber / crystal unit cell model / ensemble refinement | ||||||||||||
| Function / homology | Antifreeze, type III / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal / Antifreeze protein-like domain profile. / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal domain superfamily / extracellular region / Type-3 ice-structuring protein HPLC 12 Function and homology information Function and homology information | ||||||||||||
| Biological species |  Zoarces americanus (ocean pout) Zoarces americanus (ocean pout) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | ||||||||||||
 Authors Authors | Mikhailovskii, O. / Xue, Y. / Jia, Z. / Skrynnikov, N.R. | ||||||||||||
| Funding support |  Russian Federation, Russian Federation,  China, 3items China, 3items
| ||||||||||||
 Citation Citation |  Journal: Iucrj / Year: 2022 Journal: Iucrj / Year: 2022Title: Modeling a unit cell: crystallographic refinement procedure using the biomolecular MD simulation platform Amber. Authors: Mikhailovskii, O. / Xue, Y. / Skrynnikov, N.R. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7q3v.cif.gz 7q3v.cif.gz | 106.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7q3v.ent.gz pdb7q3v.ent.gz | 83.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7q3v.json.gz 7q3v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7q3v_validation.pdf.gz 7q3v_validation.pdf.gz | 440 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7q3v_full_validation.pdf.gz 7q3v_full_validation.pdf.gz | 440.9 KB | Display | |
| Data in XML |  7q3v_validation.xml.gz 7q3v_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  7q3v_validation.cif.gz 7q3v_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q3/7q3v https://data.pdbj.org/pub/pdb/validation_reports/q3/7q3v ftp://data.pdbj.org/pub/pdb/validation_reports/q3/7q3v ftp://data.pdbj.org/pub/pdb/validation_reports/q3/7q3v | HTTPS FTP |
-Related structure data
| Related structure data |  1msiS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 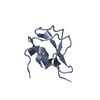
| ||||||||
| 2 | 
| ||||||||
| 3 | 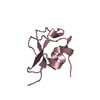
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 7056.444 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Zoarces americanus (ocean pout) / Production host: Zoarces americanus (ocean pout) / Production host:  #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.09 Å3/Da / Density % sol: 30 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 50-55% AMMONIUM SULFATE, 0.1 M SODIUM ACETATE PH 4-4.5 |
-Data collection
| Diffraction | Mean temperature: 295 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jul 11, 1996 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→14.86 Å / Num. obs: 19230 / % possible obs: 99.1 % / Redundancy: 3.8 % / Rsym value: 0.077 / Net I/σ(I): 7.8 |
| Reflection shell | Resolution: 1.9→1.97 Å / Rsym value: 0.306 / % possible all: 98.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1MSI Resolution: 1.9→14.86 Å / Cross valid method: FREE R-VALUE / Phase error: 21.18 Details: The original structure, 2MSI, has been expanded into a crystal unit cell (containing four protein chains); the resulting model was subsequently refined through a specially designed ...Details: The original structure, 2MSI, has been expanded into a crystal unit cell (containing four protein chains); the resulting model was subsequently refined through a specially designed constrained MD run using a customized version of the program Amber. Note: ordered water molecules are added after the refinement using the standard water-picking facility of phenix.refine.
| ||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||
| Displacement parameters | Biso max: 61.39 Å2 / Biso mean: 20.1134 Å2 / Biso min: 6.09 Å2 | ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→14.86 Å
| ||||||||||||||||||||
| LS refinement shell | Resolution: 1.9→1.96 Å
|
 Movie
Movie Controller
Controller



 PDBj
PDBj