[English] 日本語
 Yorodumi
Yorodumi- PDB-7nqr: Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7nqr | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z287256168 | |||||||||
 Components Components | Heat shock protein 70 | |||||||||
 Keywords Keywords | CHAPERONE / Intra-erythrocytic / AMP-PNP / Ligand | |||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of HSF1-mediated heat shock response / mRNA Splicing - Major Pathway / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / AUF1 (hnRNP D0) binds and destabilizes mRNA / Neutrophil degranulation / heat shock protein binding / protein folding chaperone / ATP-dependent protein folding chaperone / protein refolding / ATP hydrolysis activity ...Regulation of HSF1-mediated heat shock response / mRNA Splicing - Major Pathway / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / AUF1 (hnRNP D0) binds and destabilizes mRNA / Neutrophil degranulation / heat shock protein binding / protein folding chaperone / ATP-dependent protein folding chaperone / protein refolding / ATP hydrolysis activity / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.22 Å MOLECULAR REPLACEMENT / Resolution: 2.22 Å | |||||||||
 Authors Authors | Mohamad, N. / O'Donoghue, A. / Kantsadi, A.L. / Vakonakis, I. | |||||||||
| Funding support |  United Kingdom, European Union, 2items United Kingdom, European Union, 2items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands Authors: Mohamad, N. / O'Donoghue, A. / Kantsadi, A.L. / Vakonakis, I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7nqr.cif.gz 7nqr.cif.gz | 326.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7nqr.ent.gz pdb7nqr.ent.gz | 266.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7nqr.json.gz 7nqr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nq/7nqr https://data.pdbj.org/pub/pdb/validation_reports/nq/7nqr ftp://data.pdbj.org/pub/pdb/validation_reports/nq/7nqr ftp://data.pdbj.org/pub/pdb/validation_reports/nq/7nqr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7nqsC  7nquC  7nqzC  6rzqS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 42828.441 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: isolate 3D7 / Gene: PF3D7_0831700 / Plasmid: pFLOAT Details (production host): pET30a derivative, N-terminal His6-tag and HRC 3C cleavage site Production host:  |
|---|
-Non-polymers , 7 types, 239 molecules 
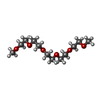
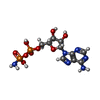


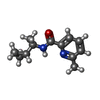







| #2: Chemical | ChemComp-GOL / #3: Chemical | ChemComp-PG6 / | #4: Chemical | #5: Chemical | #6: Chemical | #7: Chemical | ChemComp-SYQ / ( | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.45 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7.4 Details: 20 mM HEPES pH 7.4 50 mM NaCl 1 mM DTT 3.5 mM AMP-PNP 24% w/v PEG 1500 20% v/v glycerol |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.9159 Å / Beamline: I04-1 / Wavelength: 0.9159 Å |
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Jul 13, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9159 Å / Relative weight: 1 |
| Reflection | Resolution: 2.22→63.33 Å / Num. obs: 42882 / % possible obs: 100 % / Redundancy: 6.6 % / CC1/2: 1 / Rrim(I) all: 0.09 / Net I/σ(I): 11.6 |
| Reflection shell | Resolution: 2.22→2.26 Å / Redundancy: 6.8 % / Mean I/σ(I) obs: 0.9 / Num. unique obs: 2097 / CC1/2: 0.6 / Rrim(I) all: 1.931 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6RZQ Resolution: 2.22→63.33 Å / Cor.coef. Fo:Fc: 0.942 / Cor.coef. Fo:Fc free: 0.936 / SU R Cruickshank DPI: 0.278 / Cross valid method: THROUGHOUT / SU R Blow DPI: 0.283 / SU Rfree Blow DPI: 0.194 / SU Rfree Cruickshank DPI: 0.195
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 85 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.34 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.22→63.33 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.22→2.24 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







