+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7mkt | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
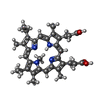
| Title | Crystal structure of r(GU)11G-NMM complex | ||||||||||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / RNA-porpyrin complex | Function / homology | : / N-METHYLMESOPORPHYRIN / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.97 Å SAD / Resolution: 1.97 Å  Authors AuthorsBingman, C.A. / Roschdi, S. / Nomura, Y. / Butcher, S.E. | Funding support | |  United States, 1items United States, 1items
 Citation Citation Journal: Nat.Struct.Mol.Biol. / Year: 2022 Journal: Nat.Struct.Mol.Biol. / Year: 2022Title: An atypical RNA quadruplex marks RNAs as vectors for gene silencing. Authors: Roschdi, S. / Yan, J. / Nomura, Y. / Escobar, C.A. / Petersen, R.J. / Bingman, C.A. / Tonelli, M. / Vivek, R. / Montemayor, E.J. / Wickens, M. / Kennedy, S.G. / Butcher, S.E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7mkt.cif.gz 7mkt.cif.gz | 33 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7mkt.ent.gz pdb7mkt.ent.gz | 22.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7mkt.json.gz 7mkt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7mkt_validation.pdf.gz 7mkt_validation.pdf.gz | 1.4 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7mkt_full_validation.pdf.gz 7mkt_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  7mkt_validation.xml.gz 7mkt_validation.xml.gz | 2.8 KB | Display | |
| Data in CIF |  7mkt_validation.cif.gz 7mkt_validation.cif.gz | 3.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mk/7mkt https://data.pdbj.org/pub/pdb/validation_reports/mk/7mkt ftp://data.pdbj.org/pub/pdb/validation_reports/mk/7mkt ftp://data.pdbj.org/pub/pdb/validation_reports/mk/7mkt | HTTPS FTP |
-Related structure data
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 7465.337 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host: synthetic construct (others) | ||
|---|---|---|---|
| #2: Chemical | ChemComp-MMP / | ||
| #3: Chemical | ChemComp-K / Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.61 Å3/Da / Density % sol: 52.9 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: The RNA smaple was prepared by mixing 1.44mM (GU)11G RNA and 1.44 mM NMM in 10 mM Tris, pH 7.03, and 100 mM KCl. An equal volume of RNA sample solution and reservoir solution were mixed and ...Details: The RNA smaple was prepared by mixing 1.44mM (GU)11G RNA and 1.44 mM NMM in 10 mM Tris, pH 7.03, and 100 mM KCl. An equal volume of RNA sample solution and reservoir solution were mixed and suspended over the reservoir solution consisting of 0.5 M Na/K tartarate, 0.1 M TrisHCl, pH 8.5, 5 mM NaOH. Cryoprotection: reservoir solution supplemented with 30% ethylene glycol. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-D / Wavelength: 1.1271 Å / Beamline: 21-ID-D / Wavelength: 1.1271 Å | |||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Sep 27, 2020 | |||||||||||||||||||||||||
| Radiation | Monochromator: Si <111> / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.1271 Å / Relative weight: 1 | |||||||||||||||||||||||||
| Reflection twin |
| |||||||||||||||||||||||||
| Reflection | Resolution: 1.97→21.61 Å / Num. obs: 5407 / % possible obs: 99.02 % / Redundancy: 6.7 % / Biso Wilson estimate: 63.54 Å2 / CC1/2: 0.998 / CC star: 1 / Rmerge(I) obs: 0.05163 / Rpim(I) all: 0.0231 / Rrim(I) all: 0.05684 / Net I/σ(I): 16.09 | |||||||||||||||||||||||||
| Reflection shell | Resolution: 1.97→2.04 Å / Redundancy: 6.9 % / Rmerge(I) obs: 0.7553 / Mean I/σ(I) obs: 1.57 / Num. unique obs: 529 / CC1/2: 0.966 / CC star: 0.991 / Rpim(I) all: 0.3083 / Rrim(I) all: 0.8172 / % possible all: 99.25 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  SAD / Resolution: 1.97→21.61 Å / Cor.coef. Fo:Fc: 0.945 / Cor.coef. Fo:Fc free: 0.944 / SU B: 3.968 / SU ML: 0.12 / Cross valid method: THROUGHOUT / ESU R: 0.051 / ESU R Free: 0.041 / Stereochemistry target values: MAXIMUM LIKELIHOOD SAD / Resolution: 1.97→21.61 Å / Cor.coef. Fo:Fc: 0.945 / Cor.coef. Fo:Fc free: 0.944 / SU B: 3.968 / SU ML: 0.12 / Cross valid method: THROUGHOUT / ESU R: 0.051 / ESU R Free: 0.041 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: Authors state that 4-way twin law was applied during refinement, while the validation program recalculated with different twin law. This causes the discrepancies between calculated and ...Details: Authors state that 4-way twin law was applied during refinement, while the validation program recalculated with different twin law. This causes the discrepancies between calculated and reported Rwork/Rfree values.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 63.361 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.97→21.61 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj