[English] 日本語
 Yorodumi
Yorodumi- PDB-7aci: In meso structure of apolipoprotein N-acyltransferase, Lnt, from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7aci | ||||||
|---|---|---|---|---|---|---|---|
| Title | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 9.8 monoacylglycerol | ||||||
 Components Components | Apolipoprotein N-acyltransferase | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / Nitrilase fold / helical bundle | ||||||
| Function / homology |  Function and homology information Function and homology informationapolipoprotein N-acyltransferase / N-acyltransferase activity / lipoprotein biosynthetic process / outer membrane-bounded periplasmic space / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Smithers, L. / van Dalsen, L. / Boland, C. / Caffrey, M. | ||||||
| Funding support |  Ireland, 1items Ireland, 1items
| ||||||
 Citation Citation |  Journal: Cryst.Growth Des. / Year: 2020 Journal: Cryst.Growth Des. / Year: 2020Title: 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins Authors: van Dalsen, L. / Smithers, L. / Boland, C. / Weichert, D. / Caffrey, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7aci.cif.gz 7aci.cif.gz | 366.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7aci.ent.gz pdb7aci.ent.gz | 251.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7aci.json.gz 7aci.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ac/7aci https://data.pdbj.org/pub/pdb/validation_reports/ac/7aci ftp://data.pdbj.org/pub/pdb/validation_reports/ac/7aci ftp://data.pdbj.org/pub/pdb/validation_reports/ac/7aci | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7acgC  5n6hS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 59280.766 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: lnt, ACU57_00505, AM464_20560, AUQ13_21565, BMA87_17500, BUE81_17200, BvCms2454_02009, BvCmsHHP001_00880, BvCmsKSNP120_02778, BvCmsKSP076_04015, BW690_14780, C5N07_14455, C9Z39_12310, CA593_ ...Gene: lnt, ACU57_00505, AM464_20560, AUQ13_21565, BMA87_17500, BUE81_17200, BvCms2454_02009, BvCmsHHP001_00880, BvCmsKSNP120_02778, BvCmsKSP076_04015, BW690_14780, C5N07_14455, C9Z39_12310, CA593_25875, CI694_20445, CIG45_13010, D0X26_17735, D2185_16075, D3821_19610, D4638_22450, D4718_15390, D9D20_13050, D9D44_16075, D9G69_11750, D9J52_13020, DBQ99_18215, DJ503_07085, DL326_16140, DT034_15925, E2119_09030, E4K55_13820, E4K60_15665, E4K61_12850, EA213_10835, EAI52_07270, EC3234A_6c00760, EC3426_01483, ECTO6_03389, ED307_12710, EEP23_08605, EI021_13075, EI028_12695, EI041_12995, EIZ93_02940, EL75_3125, EL79_3219, EL80_3175, ELT20_10985, EPT01_08450, EXX71_13910, EYD11_16175, FV293_12780, GHR40_20735, GKF74_06475, GKF86_07920, GKF89_05980, GP689_15750, GQE34_06850, GQM17_21560, GRW42_10840, NCTC8500_03946, NCTC9045_04089, NCTC9062_04723, NCTC9969_03714, PGD_02673, RK56_025370, SAMEA3472047_02047, SAMEA3472080_00307, SAMEA3484427_03116, SAMEA3484429_03266, SAMEA3752559_01009, SAMEA3753300_04108, SK85_00657 Production host:  References: UniProt: A0A037YBN4, UniProt: P23930*PLUS, apolipoprotein N-acyltransferase | ||||||
|---|---|---|---|---|---|---|---|
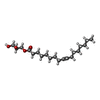
| #2: Chemical | ChemComp-GOL / #3: Chemical | ChemComp-LH9 / [( #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.45 Å3/Da / Density % sol: 49.85 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 0.1M MES pH 6.0, * %(v/v) MPD, 0.05-0.4 M sodium thiocyanate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.999 Å / Beamline: X06SA / Wavelength: 0.999 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Jun 13, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.999 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→46.57 Å / Num. obs: 26629 / % possible obs: 99.34 % / Redundancy: 12.9 % / Biso Wilson estimate: 38.94 Å2 / CC1/2: 0.99 / Net I/σ(I): 7.5 |
| Reflection shell | Resolution: 2.3→2.382 Å / Num. unique obs: 2578 / CC1/2: 0.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5N6H Resolution: 2.3→46.57 Å / SU ML: 0.2367 / Cross valid method: FREE R-VALUE / σ(F): 1.34 / Phase error: 26.3107 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→46.57 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj