+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6zpf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
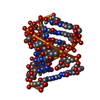
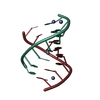
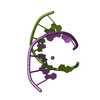
| Title | Racemic compound of RNA duplexes. | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RNA / racemic compound | |||||||||
| Function / homology | RNA Function and homology information Function and homology information | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.442 Å MOLECULAR REPLACEMENT / Resolution: 1.442 Å | |||||||||
 Authors Authors | Rypniewski, W. | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res. / Year: 2021 Journal: Nucleic Acids Res. / Year: 2021Title: Broken symmetry between RNA enantiomers in a crystal lattice. Authors: Kiliszek, A. / Blaszczyk, L. / Bejger, M. / Rypniewski, W. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6zpf.cif.gz 6zpf.cif.gz | 38.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6zpf.ent.gz pdb6zpf.ent.gz | 25.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6zpf.json.gz 6zpf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zp/6zpf https://data.pdbj.org/pub/pdb/validation_reports/zp/6zpf ftp://data.pdbj.org/pub/pdb/validation_reports/zp/6zpf ftp://data.pdbj.org/pub/pdb/validation_reports/zp/6zpf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6zq9SC  6zr1C  6zrlC  6zrsC  6zw3C  6zwuC  6zx5C  6zx8C  7a9lC  7a9nC  7a9oC  7a9pC  7a9qC  7a9rC  7a9sC  7a9tC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-RNA chain , 4 types, 4 molecules KLMN
| #1: RNA chain | Mass: 2597.601 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Chain K is D-enantiomer. It pairs with chain L. / Source: (synth.)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
|---|---|
| #2: RNA chain | Mass: 2517.553 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Chain L is D-enantiomer. It pairs with chain K. / Source: (synth.)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #3: RNA chain | Mass: 2597.601 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Chain M is L-enantiomer. It pairs with chain N. / Source: (synth.)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #4: RNA chain | Mass: 2517.553 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-Non-polymers , 2 types, 204 molecules 


| #5: Chemical | ChemComp-ZN / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.98 Å3/Da / Density % sol: 38 % / Description: needle |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: 0.2 M zinc acetate, 0.1 M cacodylate buffer, 18% w/v polyethylene glycol 8000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY PETRA III, EMBL c/o DESY  / Beamline: P13 (MX1) / Wavelength: 1.283 Å / Beamline: P13 (MX1) / Wavelength: 1.283 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Dec 1, 2018 |
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.283 Å / Relative weight: 1 |
| Reflection | Resolution: 1.44→40 Å / Num. obs: 12417 / % possible obs: 87.3 % / Redundancy: 3.5 % / CC1/2: 0.999 / Rmerge(I) obs: 0.052 / Rrim(I) all: 0.061 / Net I/σ(I): 11.04 |
| Reflection shell | Resolution: 1.44→1.53 Å / Redundancy: 3.5 % / Rmerge(I) obs: 0.293 / Mean I/σ(I) obs: 2.49 / Num. unique obs: 5931 / CC1/2: 0.963 / Rrim(I) all: 0.343 / % possible all: 74.2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6ZQ9 Resolution: 1.442→36.776 Å / Cor.coef. Fo:Fc: 0.972 / Cor.coef. Fo:Fc free: 0.958 / SU B: 1.94 / SU ML: 0.073 / Cross valid method: FREE R-VALUE / ESU R: 0.101 / ESU R Free: 0.105 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 17.245 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.442→36.776 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj
