[English] 日本語
 Yorodumi
Yorodumi- PDB-6yhw: Co-crystals in the P212121 space group, of a beta-cyclodextrin sp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6yhw | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Co-crystals in the P212121 space group, of a beta-cyclodextrin spacered by triazole heptyl from alpha-D-mannose, with FimH lectin at 2.00 A resolution. | ||||||||||||
 Components Components | FimH | ||||||||||||
 Keywords Keywords | CELL ADHESION / beta-cyclodextrin host-guest interactions inhibitor design | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpilus tip / mechanosensory behavior / cell adhesion involved in single-species biofilm formation / pilus / cell-substrate adhesion / D-mannose binding / host cell membrane / cell adhesion Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.962 Å MOLECULAR REPLACEMENT / Resolution: 1.962 Å | ||||||||||||
 Authors Authors | de Ruyck, J. / Bouckaert, J. | ||||||||||||
 Citation Citation |  Journal: Chembiochem / Year: 2016 Journal: Chembiochem / Year: 2016Title: The Antiadhesive Strategy in Crohn's Disease: Orally Active Mannosides to Decolonize Pathogenic Escherichia coli from the Gut. Authors: Alvarez Dorta, D. / Sivignon, A. / Chalopin, T. / Dumych, T.I. / Roos, G. / Bilyy, R.O. / Deniaud, D. / Krammer, E.M. / de Ruyck, J. / Lensink, M.F. / Bouckaert, J. / Barnich, N. / Gouin, S.G. #1: Journal: Chemistry / Year: 2013 Title: Heptyl a-D-mannosides grafted on a b-cyclodextrin core to interfere with Escherichia coli adhesion: an in vivo multivalent effect. Authors: Bouckaert, J. / Li, Z. / Xavier, C. / Almant, M. / Caveliers, V. / Lahoutte, T. / Weeks, S.D. / Kovensky, J. / Gouin, S.G. #2:  Journal: Chembiochem / Year: 2016 Journal: Chembiochem / Year: 2016Title: The Antiadhesive Strategy in Crohn's Disease: Orally Active Mannosides to Decolonize Pathogenic Escherichia coli from the Gut. Authors: Alvarez Dorta, D. / Sivignon, A. / Chalopin, T. / Dumych, T.I. / Roos, G. / Bilyy, R.O. / Deniaud, D. / Krammer, E.M. / de Ruyck, J. / Lensink, M.F. / Bouckaert, J. / Barnich, N. / Gouin, S.G. #3: Journal: Molecules / Year: 2018 Title: Targeting Dynamical Binding Processes in the Design of Non-Antibiotic Anti-Adhesives by Molecular Simulation-The Example of FimH. Authors: Krammer, E.M. / de Ruyck, J. / Roos, G. / Bouckaert, J. / Lensink, M.F. #4: Journal: Expert Opin Ther Targets / Year: 2017 Title: The potential of FimH as a novel therapeutic target for the treatment of Crohn's disease. Authors: Sivignon, A. / Bouckaert, J. / Bernard, J. / Gouin, S.G. / Barnich, N. #5:  Book title: Biophysical Techniques in Drug Discovery / Journal: Biophysical Techniques in Drug Discovery / Year: 2018 Book title: Biophysical Techniques in Drug Discovery / Journal: Biophysical Techniques in Drug Discovery / Year: 2018Title: Chapter 4 Molecular Mechanisms of Drug Action: X-ray Crystallography at the Basis of Structure-based and Ligand-based Drug Design Authors: de Ruyck, J. / Roos, G. / Krammer, E.-M. / Prevost, M. / Lensink, M.F. / Bouckaert, J. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6yhw.cif.gz 6yhw.cif.gz | 151.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6yhw.ent.gz pdb6yhw.ent.gz | 117.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6yhw.json.gz 6yhw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yh/6yhw https://data.pdbj.org/pub/pdb/validation_reports/yh/6yhw ftp://data.pdbj.org/pub/pdb/validation_reports/yh/6yhw ftp://data.pdbj.org/pub/pdb/validation_reports/yh/6yhw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1uwfS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 31488.260 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: fimH, fimH_1, fimH_2, fimH_3, sfaH_2, A6581_19835, ACN81_00940, AKG99_10920, AM446_24165, AM464_16260, AUQ13_11280, AWG78_008695, B6V57_25500, B9M99_24715, B9T59_08965, BON65_08035, BON69_ ...Gene: fimH, fimH_1, fimH_2, fimH_3, sfaH_2, A6581_19835, ACN81_00940, AKG99_10920, AM446_24165, AM464_16260, AUQ13_11280, AWG78_008695, B6V57_25500, B9M99_24715, B9T59_08965, BON65_08035, BON69_13145, BON76_10045, BON87_19590, BvCms12BK_05011, BvCms2454_04206, C2U48_10910, C5P44_07835, C6B13_05760, CR538_22045, CRX46_08755, D2188_19245, D9G11_20470, D9H70_11375, D9I97_23340, D9J60_03345, DJ503_16470, DNQ45_27015, DS732_03965, DU321_25405, DXT73_23425, E0L12_19135, EC3234A_192c00060, ECONIH1_25860, ECTO6_04155, EIA21_01475, ELV08_23920, EQ830_16720, ERS085383_04267, F0312_06085, F1E19_12715, FNJ69_21090, FNJ83_00985, FV293_11400, FY127_19655, MS6198_50840, MS8345_04977, NCTC10090_02366, NCTC8450_00821, NCTC9045_05036, SAMEA3472056_01232, YDC107_3079 Production host:  |
|---|
-Sugars , 2 types, 4 molecules 
| #2: Polysaccharide | | #5: Sugar | |
|---|
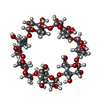
-Non-polymers , 3 types, 354 molecules 




| #3: Chemical | | #4: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.64 Å3/Da / Density % sol: 24.9 % / Description: large bars |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: FimH (10 mg/mL) with beta-cyclodextrin triazole heptyl a-d-mannose (2 mM). Crystals were equilibrated against a well containing sodium citrate tribasic dihydrate buffer (pH 6.5, 1.6M). Prior ...Details: FimH (10 mg/mL) with beta-cyclodextrin triazole heptyl a-d-mannose (2 mM). Crystals were equilibrated against a well containing sodium citrate tribasic dihydrate buffer (pH 6.5, 1.6M). Prior to data collection, crystals were cryoprotected with use of reservoir solution containing glycerol (20%) and flash-cooled in liquid nitrogen. |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID29 / Wavelength: 1 Å / Beamline: ID29 / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Nov 18, 2015 |
| Radiation | Monochromator: liquid nitrogen cooled channel-cut silicon monochromator Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.962→48.348 Å / Num. obs: 29988 / % possible obs: 98.92 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.3 % / Biso Wilson estimate: 21.38 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 7.6 |
| Reflection shell | Resolution: 1.962→2.161 Å / Rmerge(I) obs: 0.667 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 7379 / % possible all: 98.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1UWF Resolution: 1.962→46.348 Å / Cross valid method: FREE R-VALUE / σ(F): 0 / Phase error: 29.91
| ||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.12 Å2 | ||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.23 Å | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.962→46.348 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.962→2.025 Å
|
 Movie
Movie Controller
Controller










 PDBj
PDBj