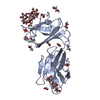
Entry Database : PDB / ID : 6y94Title Ca2+-bound Calmodulin mutant N53I Calmodulin Keywords / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / Authors Holt, C. / Nielsen, L.H. / Lau, K. / Brohus, M. / Sorensen, A.B. / Larsen, K.T. / Sommer, C. / Petegem, F.V. / Overgaard, M.T. / Wimmer, R. Funding support Organization Grant number Country Danish Council for Independent Research DFF-1323-00344 Novo Nordisk Foundation NNF18OC0053032 The Carlsberg Foundation European Union (EU) 261863 European Union The SparNord foundation The Obel Family foundation
Journal : J.Biol.Chem. / Year : 2020Title : The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.Authors : Holt, C. / Hamborg, L. / Lau, K. / Brohus, M. / Sorensen, A.B. / Larsen, K.T. / Sommer, C. / Van Petegem, F. / Overgaard, M.T. / Wimmer, R. History Deposition Mar 6, 2020 Deposition site / Processing site Revision 1.0 Apr 29, 2020 Provider / Type Revision 1.1 May 6, 2020 Group / Category / citation_authorItem _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed ... _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID / _citation_author.name Revision 1.2 Jun 10, 2020 Group / Category / citation_authorItem _citation.journal_volume / _citation.page_first ... _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.title / _citation_author.identifier_ORCID Revision 1.3 Jun 14, 2023 Group / Other / Category / pdbx_database_statusItem / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_nmr_dataRevision 1.4 Jun 19, 2024 Group / Database references / Category / chem_comp_bond / database_2 / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Denmark, European Union, 6items
Denmark, European Union, 6items  Citation
Citation Journal: J.Biol.Chem. / Year: 2020
Journal: J.Biol.Chem. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6y94.cif.gz
6y94.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6y94.ent.gz
pdb6y94.ent.gz PDB format
PDB format 6y94.json.gz
6y94.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/y9/6y94
https://data.pdbj.org/pub/pdb/validation_reports/y9/6y94 ftp://data.pdbj.org/pub/pdb/validation_reports/y9/6y94
ftp://data.pdbj.org/pub/pdb/validation_reports/y9/6y94


 Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: CALM1, CALM, CAM, CAM1 / Production host:
Homo sapiens (human) / Gene: CALM1, CALM, CAM, CAM1 / Production host: 
 Sample preparation
Sample preparation Movie
Movie Controller
Controller












 PDBj
PDBj






















 HSQC
HSQC