[English] 日本語
 Yorodumi
Yorodumi- PDB-6x3x: Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6x3x | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus diazepam | ||||||||||||||||||
 Components Components |
| ||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Ion channel / Cys-loop receptor / pentametic ligand gated channel / GABAA receptor | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationbenzodiazepine receptor activity / GABA receptor complex / inner ear receptor cell development / cellular response to histamine / GABA receptor activation / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / inhibitory synapse assembly / innervation ...benzodiazepine receptor activity / GABA receptor complex / inner ear receptor cell development / cellular response to histamine / GABA receptor activation / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / inhibitory synapse assembly / innervation / synaptic transmission, GABAergic / gamma-aminobutyric acid signaling pathway / postsynaptic specialization membrane / neurotransmitter receptor activity / chloride channel activity / cochlea development / adult behavior / Signaling by ERBB4 / chloride channel complex / regulation of postsynaptic membrane potential / transmembrane transporter complex / GABA-ergic synapse / chloride transmembrane transport / dendrite membrane / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / cytoplasmic vesicle membrane / chemical synaptic transmission / postsynaptic membrane / postsynapse / dendritic spine / neuron projection / axon / synapse / extracellular exosome / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.92 Å | ||||||||||||||||||
 Authors Authors | Kim, J.J. / Gharpure, A. / Teng, J. / Zhuang, Y. / Howard, R.J. / Zhu, S. / Noviello, C.M. / Walsh, R.M. / Lindahl, E. / Hibbs, R.E. | ||||||||||||||||||
| Funding support |  United States, 5items United States, 5items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Shared structural mechanisms of general anaesthetics and benzodiazepines. Authors: Jeong Joo Kim / Anant Gharpure / Jinfeng Teng / Yuxuan Zhuang / Rebecca J Howard / Shaotong Zhu / Colleen M Noviello / Richard M Walsh / Erik Lindahl / Ryan E Hibbs /   Abstract: Most general anaesthetics and classical benzodiazepine drugs act through positive modulation of γ-aminobutyric acid type A (GABA) receptors to dampen neuronal activity in the brain. However, direct ...Most general anaesthetics and classical benzodiazepine drugs act through positive modulation of γ-aminobutyric acid type A (GABA) receptors to dampen neuronal activity in the brain. However, direct structural information on the mechanisms of general anaesthetics at their physiological receptor sites is lacking. Here we present cryo-electron microscopy structures of GABA receptors bound to intravenous anaesthetics, benzodiazepines and inhibitory modulators. These structures were solved in a lipidic environment and are complemented by electrophysiology and molecular dynamics simulations. Structures of GABA receptors in complex with the anaesthetics phenobarbital, etomidate and propofol reveal both distinct and common transmembrane binding sites, which are shared in part by the benzodiazepine drug diazepam. Structures in which GABA receptors are bound by benzodiazepine-site ligands identify an additional membrane binding site for diazepam and suggest an allosteric mechanism for anaesthetic reversal by flumazenil. This study provides a foundation for understanding how pharmacologically diverse and clinically essential drugs act through overlapping and distinct mechanisms to potentiate inhibitory signalling in the brain. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6x3x.cif.gz 6x3x.cif.gz | 409.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6x3x.ent.gz pdb6x3x.ent.gz | 322.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6x3x.json.gz 6x3x.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x3/6x3x https://data.pdbj.org/pub/pdb/validation_reports/x3/6x3x ftp://data.pdbj.org/pub/pdb/validation_reports/x3/6x3x ftp://data.pdbj.org/pub/pdb/validation_reports/x3/6x3x | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  22036MC  6x3sC  6x3tC  6x3uC  6x3vC  6x3wC  6x3zC  6x40C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Gamma-aminobutyric acid receptor subunit ... , 3 types, 5 molecules ACBDE
| #1: Protein | Mass: 41810.086 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABRB2 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host: Homo sapiens (human) / Gene: GABRB2 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host:  Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P47870 Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P47870#2: Protein | Mass: 41061.211 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABRA1 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host: Homo sapiens (human) / Gene: GABRA1 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host:  Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P14867 Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P14867#3: Protein | | Mass: 47673.109 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABRG2 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host: Homo sapiens (human) / Gene: GABRG2 / Plasmid: pEZT-BM / Cell line (production host): HEK293S / Production host:  Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P18507 Homo sapiens (human) / Variant (production host): GnTI- / References: UniProt: P18507 |
|---|
-Antibody , 2 types, 4 molecules ILJK
| #4: Antibody | Mass: 23505.943 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  #5: Antibody | Mass: 49811.043 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Sugars , 4 types, 7 molecules 
| #6: Polysaccharide | Source method: isolated from a genetically manipulated source #7: Polysaccharide | alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D- ...alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source #8: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source #9: Sugar | |
|---|
-Non-polymers , 2 types, 6 molecules 
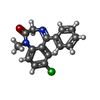

| #10: Chemical | | #11: Chemical | ChemComp-DZP / |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| ||||||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: HEK293T GnT1- Homo sapiens (human) / Cell: HEK293T GnT1- | ||||||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | ||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||
| Specimen | Conc.: 7.7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: 3.5 second blot |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 63.04 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of real images: 11213 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.16_3549: / Classification: refinement | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2868814 | ||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.92 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 297028 / Algorithm: BACK PROJECTION / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL | ||||||||||||||||||||||||||||||||||||
| Refinement | Highest resolution: 2.92 Å | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller


















 PDBj
PDBj







