[English] 日本語
 Yorodumi
Yorodumi- PDB-6tjt: Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6tjt | ||||||
|---|---|---|---|---|---|---|---|
| Title | Neuropilin2-b1 domain in a complex with the C-terminal VEGFC peptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / VEGF-binding / NRP2 / angiogenesis / immunomodulation | ||||||
| Function / homology |  Function and homology information Function and homology informationvascular endothelial growth factor receptor 3 binding / substrate-dependent cell migration / vestibulocochlear nerve structural organization / dorsal root ganglion morphogenesis / ventral trunk neural crest cell migration / sympathetic neuron projection guidance / facioacoustic ganglion development / trigeminal ganglion development / trigeminal nerve structural organization / sensory neuron axon guidance ...vascular endothelial growth factor receptor 3 binding / substrate-dependent cell migration / vestibulocochlear nerve structural organization / dorsal root ganglion morphogenesis / ventral trunk neural crest cell migration / sympathetic neuron projection guidance / facioacoustic ganglion development / trigeminal ganglion development / trigeminal nerve structural organization / sensory neuron axon guidance / positive regulation of lymphangiogenesis / facial nerve structural organization / branchiomotor neuron axon guidance / gonadotrophin-releasing hormone neuronal migration to the hypothalamus / VEGF ligand-receptor interactions / positive regulation of mast cell chemotaxis / axon extension involved in axon guidance / morphogenesis of embryonic epithelium / sympathetic neuron projection extension / NrCAM interactions / Neurophilin interactions with VEGF and VEGFR / regulation of vascular endothelial growth factor receptor signaling pathway / positive regulation of mesenchymal stem cell proliferation / VEGF binds to VEGFR leading to receptor dimerization / neural crest cell migration involved in autonomic nervous system development / sympathetic ganglion development / vascular endothelial growth factor receptor activity / nerve development / semaphorin receptor complex / induction of positive chemotaxis / semaphorin receptor activity / outflow tract septum morphogenesis / sprouting angiogenesis / regulation of postsynapse organization / vascular endothelial growth factor signaling pathway / negative chemotaxis / growth factor binding / positive regulation of neuroblast proliferation / cytokine binding / chemoattractant activity / semaphorin-plexin signaling pathway / positive regulation of cell division / positive regulation of blood vessel endothelial cell migration / positive regulation of glial cell proliferation / vascular endothelial growth factor receptor signaling pathway / negative regulation of osteoblast differentiation / glial cell proliferation / negative regulation of blood pressure / positive regulation of endothelial cell proliferation / positive regulation of endothelial cell migration / axon guidance / platelet alpha granule lumen / animal organ morphogenesis / positive regulation of epithelial cell proliferation / cellular response to leukemia inhibitory factor / positive regulation of protein secretion / growth factor activity / positive regulation of angiogenesis / Platelet degranulation / signaling receptor activity / heparin binding / angiogenesis / response to hypoxia / postsynaptic membrane / cell adhesion / response to xenobiotic stimulus / axon / positive regulation of cell population proliferation / glutamatergic synapse / signal transduction / extracellular space / extracellular region / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.31 Å MOLECULAR REPLACEMENT / Resolution: 1.31 Å | ||||||
 Authors Authors | Eldrid, C. / Yu, L. / Yelland, T. / Fotinou, C. / Djordjevic, S. | ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: NRP2-b1 domain in a complex with the C-terminal VEGFC peptide Authors: Eldrid, C. / Yu, L. / Yelland, T. / Fotinou, C. / Djordjevic, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6tjt.cif.gz 6tjt.cif.gz | 185.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6tjt.ent.gz pdb6tjt.ent.gz | 121.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6tjt.json.gz 6tjt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tj/6tjt https://data.pdbj.org/pub/pdb/validation_reports/tj/6tjt ftp://data.pdbj.org/pub/pdb/validation_reports/tj/6tjt ftp://data.pdbj.org/pub/pdb/validation_reports/tj/6tjt | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4rn5S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 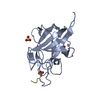
| ||||||||||||
| 2 | 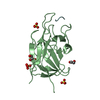
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 18368.584 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: The first N-terminal residue and the last two C-terminal residues from the protein construct were not observed in the electron density maps. Source: (gene. exp.)  Homo sapiens (human) / Gene: NRP2, VEGF165R2 / Production host: Homo sapiens (human) / Gene: NRP2, VEGF165R2 / Production host:  #2: Protein/peptide | Mass: 671.834 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: The electron density for the acetylated N-terminal Ser and the two Ile residues did not allow for the modeling of these residues. Source: (synth.)  Homo sapiens (human) / References: UniProt: P49767*PLUS Homo sapiens (human) / References: UniProt: P49767*PLUS#3: Chemical | ChemComp-SO4 / #4: Chemical | #5: Water | ChemComp-HOH / | Has ligand of interest | N | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 49.2 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion Details: 0.1 M NaOAc, 0.1 M HEPES pH 7.5, 12 % (w/v) PEG4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I03 / Wavelength: 0.976 Å / Beamline: I03 / Wavelength: 0.976 Å | ||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Jan 23, 2016 | ||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength | Wavelength: 0.976 Å / Relative weight: 1 | ||||||||||||
| Reflection | Resolution: 1.31→63 Å / Num. obs: 86786 / % possible obs: 98.9 % / Redundancy: 3.1 % / Biso Wilson estimate: 13.52 Å2 / CC1/2: 0.998 / Net I/σ(I): 12.6 | ||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4RN5 Resolution: 1.31→48.64 Å / SU ML: 0.1122 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 20.0195
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.74 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.31→48.64 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj








