[English] 日本語
 Yorodumi
Yorodumi- PDB-6qgr: The F420-reducing [NiFe] hydrogenase complex from Methanosarcina ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6qgr | ||||||
|---|---|---|---|---|---|---|---|
| Title | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | ||||||
 Components Components | (Coenzyme F420 hydrogenase subunit ...) x 3 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / [NiFe] containing hydrogenase / reversible oxidation of dihydrogen / oxygen sensitive / methanogenic acetoclastic Archaea / METAL BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationcoenzyme F420 hydrogenase / coenzyme F420 hydrogenase activity / oxidoreductase activity, acting on CH or CH2 groups, with an iron-sulfur protein as acceptor / ferredoxin hydrogenase activity / nickel cation binding / 2 iron, 2 sulfur cluster binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding Similarity search - Function | ||||||
| Biological species |  Methanosarcina barkeri MS (archaea) Methanosarcina barkeri MS (archaea) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.839 Å MOLECULAR REPLACEMENT / Resolution: 1.839 Å | ||||||
 Authors Authors | Ilina, Y. / Lorent, C. / Katz, S. / Jeoung, J.H. / Shima, S. / Horch, M. / Zebger, I. / Dobbek, H. | ||||||
| Funding support |  Germany, 1items Germany, 1items
| ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2019 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2019Title: X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases. Authors: Ilina, Y. / Lorent, C. / Katz, S. / Jeoung, J.H. / Shima, S. / Horch, M. / Zebger, I. / Dobbek, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6qgr.cif.gz 6qgr.cif.gz | 235.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6qgr.ent.gz pdb6qgr.ent.gz | 180.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6qgr.json.gz 6qgr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qg/6qgr https://data.pdbj.org/pub/pdb/validation_reports/qg/6qgr ftp://data.pdbj.org/pub/pdb/validation_reports/qg/6qgr ftp://data.pdbj.org/pub/pdb/validation_reports/qg/6qgr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6qgtC  6qiiC  4omfS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | x 12
| |||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Coenzyme F420 hydrogenase subunit ... , 3 types, 3 molecules GAB
| #1: Protein | Mass: 27598.912 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3LP72, coenzyme F420 hydrogenase Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3LP72, coenzyme F420 hydrogenase |
|---|---|
| #2: Protein | Mass: 48531.621 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: FRH-A harbours an active site for the dihydrogen catalysis Source: (natural)  Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3QYL7*PLUS Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3QYL7*PLUS |
| #3: Protein | Mass: 32297.244 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3QWH3, coenzyme F420 hydrogenase Methanosarcina barkeri MS (archaea) / References: UniProt: A0A0E3QWH3, coenzyme F420 hydrogenase |
-Non-polymers , 10 types, 670 molecules 

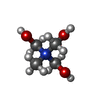
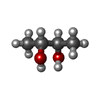

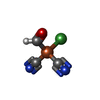








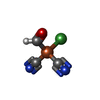




| #4: Chemical | ChemComp-SF4 / #5: Chemical | ChemComp-FES / | #6: Chemical | #7: Chemical | ChemComp-BU3 / ( #8: Chemical | ChemComp-MPD / ( #9: Chemical | ChemComp-NFU / | #10: Chemical | ChemComp-FE / | #11: Chemical | ChemComp-MG / | #12: Chemical | ChemComp-FAD / | #13: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.31 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / Details: Tris, MPD, PEG1000 |
-Data collection
| Diffraction | Mean temperature: 80 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.981 Å / Beamline: 14.1 / Wavelength: 0.981 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 25, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.981 Å / Relative weight: 1 |
| Reflection | Resolution: 1.839→50 Å / Num. obs: 183344 / % possible obs: 99.57 % / Redundancy: 4 % / Rrim(I) all: 0.211 / Net I/σ(I): 6.62 |
| Reflection shell | Resolution: 1.8395→1.8823 Å |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4OMF Resolution: 1.839→48.138 Å / SU ML: 0.39 / Cross valid method: FREE R-VALUE / σ(F): 0.78 / Phase error: 28.01
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.839→48.138 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller








 PDBj
PDBj




















