| Entry | Database: PDB / ID: 6q3w
|
|---|
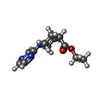
| Title | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate |
|---|
 Components Components | Galactokinase |
|---|
 Keywords Keywords | TRANSFERASE / GALK1 / fragment / fragment-bound / galactokinase 1 / sugar kinase |
|---|
| Function / homology |  Function and homology information Function and homology information
glycolytic process from galactose / Defective GALK1 causes GALCT2 / galactitol metabolic process / galactokinase / galactokinase activity / galactose catabolic process via UDP-galactose, Leloir pathway / galactose binding / galactose metabolic process / Galactose catabolism / extracellular exosome ...glycolytic process from galactose / Defective GALK1 causes GALCT2 / galactitol metabolic process / galactokinase / galactokinase activity / galactose catabolic process via UDP-galactose, Leloir pathway / galactose binding / galactose metabolic process / Galactose catabolism / extracellular exosome / ATP binding / membrane / cytoplasm / cytosolSimilarity search - Function Galactokinase, N-terminal domain / Galactokinase, conserved site / Galactokinase galactose-binding signature / Galactokinase signature. / Galactokinase / Mevalonate/galactokinase / GHMP kinase, ATP-binding, conserved site / GHMP kinases putative ATP-binding domain. / GHMP kinase, C-terminal domain / GHMP kinases C terminal ...Galactokinase, N-terminal domain / Galactokinase, conserved site / Galactokinase galactose-binding signature / Galactokinase signature. / Galactokinase / Mevalonate/galactokinase / GHMP kinase, ATP-binding, conserved site / GHMP kinases putative ATP-binding domain. / GHMP kinase, C-terminal domain / GHMP kinases C terminal / GHMP kinase, C-terminal domain / GHMP kinase N-terminal domain / GHMP kinases N terminal domain / GHMP kinase, C-terminal domain superfamily / Ribosomal Protein S5; domain 2 - #10 / Ribosomal Protein S5; domain 2 / Ribosomal protein S5 domain 2-type fold, subgroup / Ribosomal protein S5 domain 2-type fold / Alpha-Beta Plaits / 2-Layer Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.962 Å MOLECULAR REPLACEMENT / Resolution: 1.962 Å |
|---|
 Authors Authors | Mackinnon, S.R. / Bezerra, G.A. / Zhang, M. / Foster, W. / Krojer, T. / Brandao-Neto, J. / Douangamath, A. / Arrowsmith, C. / Edwards, A. / Bountra, C. ...Mackinnon, S.R. / Bezerra, G.A. / Zhang, M. / Foster, W. / Krojer, T. / Brandao-Neto, J. / Douangamath, A. / Arrowsmith, C. / Edwards, A. / Bountra, C. / Brennan, P. / Lai, K. / Yue, W.W. |
|---|
| Funding support |  United Kingdom, 1items United Kingdom, 1items | Organization | Grant number | Country |
|---|
| Wellcome Trust | 092809/Z/10/Z |  United Kingdom United Kingdom |
|
|---|
 Citation Citation |  Journal: To Be Published Journal: To Be Published
Title: Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate
Authors: Mackinnon, S.R. / Bezerra, G.A. / Zhang, M. / Foster, W. / Krojer, T. / Brandao-Neto, J. / Douangamath, A. / Arrowsmith, C. / Edwards, A. / Bountra, C. / Brennan, P. / Lai, K. / Yue, W.W. |
|---|
| History | | Deposition | Dec 4, 2018 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Jan 23, 2019 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 29, 2020 | Group: Data collection / Derived calculations / Structure summary
Category: chem_comp / entity ...chem_comp / entity / pdbx_chem_comp_identifier / pdbx_entity_nonpoly / struct_conn / struct_site / struct_site_gen
Item: _chem_comp.name / _chem_comp.type ..._chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_symmetry
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 1.2 | Jan 24, 2024 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 1.3 | Oct 16, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.962 Å
MOLECULAR REPLACEMENT / Resolution: 1.962 Å  Authors
Authors United Kingdom, 1items
United Kingdom, 1items  Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6q3w.cif.gz
6q3w.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6q3w.ent.gz
pdb6q3w.ent.gz PDB format
PDB format 6q3w.json.gz
6q3w.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/q3/6q3w
https://data.pdbj.org/pub/pdb/validation_reports/q3/6q3w ftp://data.pdbj.org/pub/pdb/validation_reports/q3/6q3w
ftp://data.pdbj.org/pub/pdb/validation_reports/q3/6q3w
 Links
Links Assembly
Assembly




 Components
Components Homo sapiens (human) / Gene: GALK1, GALK / Plasmid: pNIC24-Bsa / Production host:
Homo sapiens (human) / Gene: GALK1, GALK / Plasmid: pNIC24-Bsa / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I04-1 / Wavelength: 0.91587 Å
/ Beamline: I04-1 / Wavelength: 0.91587 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj