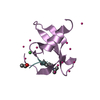
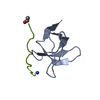
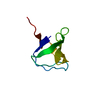
Entry Database : PDB / ID : 6mwmTitle Bat coronavirus HKU4 SUD-C Non-structural protein 3 Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Method / Authors Staup, A.J. / De Silva, I.U. / Catt, J.T. / Tan, X. / Hammond, R.G. / Johnson, M.A. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R35GM119456
Journal : Nat Prod Commun / Year : 2019Title : Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.Authors : Staup, A.J. / De Silva, I.U. / Catt, J.T. / Tan, X. / Hammond, R.G. / Johnson, M.A. History Deposition Oct 29, 2018 Deposition site / Processing site Revision 1.0 Sep 11, 2019 Provider / Type Revision 1.1 Jan 1, 2020 Group / Database references / Category / pdbx_audit_supportItem / _citation.journal_id_ISSN / _pdbx_audit_support.funding_organizationRevision 1.2 May 27, 2020 Group / Category / citation_authorItem _citation.journal_id_ISSN / _citation.journal_volume ... _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID Revision 1.3 Aug 26, 2020 Group / Category Revision 1.4 Jun 14, 2023 Group / Other / Category / pdbx_database_statusItem / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_nmr_dataRevision 1.5 May 15, 2024 Group / Database references / Category / chem_comp_bond / database_2 / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Bat coronavirus HKU4
Bat coronavirus HKU4 Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Nat Prod Commun / Year: 2019
Journal: Nat Prod Commun / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6mwm.cif.gz
6mwm.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6mwm.ent.gz
pdb6mwm.ent.gz PDB format
PDB format 6mwm.json.gz
6mwm.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/mw/6mwm
https://data.pdbj.org/pub/pdb/validation_reports/mw/6mwm ftp://data.pdbj.org/pub/pdb/validation_reports/mw/6mwm
ftp://data.pdbj.org/pub/pdb/validation_reports/mw/6mwm Links
Links Assembly
Assembly
 Components
Components Bat coronavirus HKU4 / Gene: rep, 1a-1b / Plasmid: pET15b-TEV / Production host:
Bat coronavirus HKU4 / Gene: rep, 1a-1b / Plasmid: pET15b-TEV / Production host: 
 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller










 PDBj
PDBj





 gel filtration
gel filtration