+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6lw0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | ||||||
 Components Components | Toll-like receptor 7 | ||||||
 Keywords Keywords | IMMUNE SYSTEM / TLR7 / antagonist | ||||||
| Function / homology |  Function and homology information Function and homology informationtoll-like receptor 7 signaling pathway / toll-like receptor 8 signaling pathway / response to cGMP / early phagosome / siRNA binding / endolysosome membrane / positive regulation of macrophage cytokine production / toll-like receptor signaling pathway / pattern recognition receptor activity / positive regulation of interferon-alpha production ...toll-like receptor 7 signaling pathway / toll-like receptor 8 signaling pathway / response to cGMP / early phagosome / siRNA binding / endolysosome membrane / positive regulation of macrophage cytokine production / toll-like receptor signaling pathway / pattern recognition receptor activity / positive regulation of interferon-alpha production / canonical NF-kappaB signal transduction / positive regulation of chemokine production / JNK cascade / positive regulation of interferon-beta production / positive regulation of interleukin-8 production / cellular response to mechanical stimulus / cellular response to virus / positive regulation of interleukin-6 production / positive regulation of type II interferon production / positive regulation of inflammatory response / double-stranded RNA binding / defense response to virus / positive regulation of canonical NF-kappaB signal transduction / receptor complex / single-stranded RNA binding / inflammatory response / innate immune response / endoplasmic reticulum / positive regulation of transcription by RNA polymerase II / metal ion binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | ||||||
 Authors Authors | Zhang, Z. / Ohto, U. / Shimizu, T. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structural analysis reveals TLR7 dynamics underlying antagonism. Authors: Shingo Tojo / Zhikuan Zhang / Hiroyuki Matsui / Masahiro Tahara / Mitsunori Ikeguchi / Mami Kochi / Mami Kamada / Hideki Shigematsu / Akihisa Tsutsumi / Naruhiko Adachi / Takuma Shibata / ...Authors: Shingo Tojo / Zhikuan Zhang / Hiroyuki Matsui / Masahiro Tahara / Mitsunori Ikeguchi / Mami Kochi / Mami Kamada / Hideki Shigematsu / Akihisa Tsutsumi / Naruhiko Adachi / Takuma Shibata / Masaki Yamamoto / Masahide Kikkawa / Toshiya Senda / Yoshiaki Isobe / Umeharu Ohto / Toshiyuki Shimizu /  Abstract: Toll-like receptor 7 (TLR7) recognizes both microbial and endogenous RNAs and nucleosides. Aberrant activation of TLR7 has been implicated in several autoimmune diseases including systemic lupus ...Toll-like receptor 7 (TLR7) recognizes both microbial and endogenous RNAs and nucleosides. Aberrant activation of TLR7 has been implicated in several autoimmune diseases including systemic lupus erythematosus (SLE). Here, by modifying potent TLR7 agonists, we develop a series of TLR7-specific antagonists as promising therapeutic agents for SLE. These compounds protect mice against lethal autoimmunity. Combining crystallography and cryo-electron microscopy, we identify the open conformation of the receptor and reveal the structural equilibrium between open and closed conformations that underlies TLR7 antagonism, as well as the detailed mechanism by which TLR7-specific antagonists bind to their binding pocket in TLR7. Our work provides small-molecule TLR7-specific antagonists and suggests the TLR7-targeting strategy for treating autoimmune diseases. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6lw0.cif.gz 6lw0.cif.gz | 328.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6lw0.ent.gz pdb6lw0.ent.gz | 261.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6lw0.json.gz 6lw0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lw/6lw0 https://data.pdbj.org/pub/pdb/validation_reports/lw/6lw0 ftp://data.pdbj.org/pub/pdb/validation_reports/lw/6lw0 ftp://data.pdbj.org/pub/pdb/validation_reports/lw/6lw0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  0999C  1000C  6lvxC  6lvyC  6lvzC  6lw1C  5gmhS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: ALA / Beg label comp-ID: ALA / End auth comp-ID: LEU / End label comp-ID: LEU / Auth seq-ID: 27 - 835 / Label seq-ID: 5 - 813
|
- Components
Components
-Protein , 1 types, 2 molecules BA
| #1: Protein | Mass: 94745.594 Da / Num. of mol.: 2 Mutation: N145Q/N367Q/S418L/E419VV420P/G421R/F422G/C423S/N466Q/N777Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Sugars , 2 types, 13 molecules 
| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Sugar | ChemComp-NAG / |
|---|
-Non-polymers , 3 types, 32 molecules 
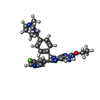



| #4: Chemical | ChemComp-SO4 / #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.69 Å3/Da / Density % sol: 54.26 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.2M ammonium sulfate, 0.1M tri-sodium citrate pH 5.6, 15% (w/v) PEG 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NE3A / Wavelength: 1 Å / Beamline: AR-NE3A / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: May 14, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→46.78 Å / Num. obs: 63582 / % possible obs: 100 % / Redundancy: 6.7 % / CC1/2: 0.998 / Net I/σ(I): 13.8 |
| Reflection shell | Resolution: 2.6→2.66 Å / Num. unique obs: 4430 / CC1/2: 0.612 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5GMH Resolution: 2.6→46.291 Å / SU ML: 0.39 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 27.8
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 118.42 Å2 / Biso mean: 56.3342 Å2 / Biso min: 24.13 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.6→46.291 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / % reflection obs: 100 %
|
 Movie
Movie Controller
Controller

















 PDBj
PDBj







