Entry Database : PDB / ID : 6bz3Title Complex structure of FAK FAT domain and DCC P3 motif Focal adhesion kinase 1 Netrin receptor DCC Keywords / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Rattus norvegicus (Norway rat)Method / / / Resolution : 2.497 Å Authors Xu, S. / Wang, J.-H. Funding support Organization Grant number Country National Institutes of Health/National Heart, Lung, and Blood Institute (NIH/NHLBI) P01HL103526
Journal : Cell Discov / Year : 2018Title : The binding of DCC-P3 motif and FAK-FAT domain mediates the initial step of netrin-1/DCC signaling for axon attraction.Authors : Xu, S. / Liu, Y. / Li, X. / Liu, Y. / Meijers, R. / Zhang, Y. / Wang, J.H. History Deposition Dec 22, 2017 Deposition site / Processing site Revision 1.0 Apr 4, 2018 Provider / Type Revision 1.1 Dec 4, 2019 Group / Category / Item Revision 1.2 Oct 4, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information

 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.497 Å
MOLECULAR REPLACEMENT / Resolution: 2.497 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Cell Discov / Year: 2018
Journal: Cell Discov / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6bz3.cif.gz
6bz3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6bz3.ent.gz
pdb6bz3.ent.gz PDB format
PDB format 6bz3.json.gz
6bz3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/bz/6bz3
https://data.pdbj.org/pub/pdb/validation_reports/bz/6bz3 ftp://data.pdbj.org/pub/pdb/validation_reports/bz/6bz3
ftp://data.pdbj.org/pub/pdb/validation_reports/bz/6bz3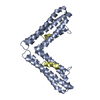
 Links
Links Assembly
Assembly
 Components
Components


 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 19-ID / Wavelength: 0.979 Å
/ Beamline: 19-ID / Wavelength: 0.979 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj















