[English] 日本語
 Yorodumi
Yorodumi- PDB-5xv9: Solution Structure of Cold Shock Protein from Colwellia psychrery... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5xv9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution Structure of Cold Shock Protein from Colwellia psychrerythraea | ||||||
 Components Components | Cold-shock DNA-binding domain family protein | ||||||
 Keywords Keywords | RNA BINDING PROTEIN / Cold-shock protein / Psychrophile / NMR spectroscopy / solution structure | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Colwellia psychrerythraea (bacteria) Colwellia psychrerythraea (bacteria) | ||||||
| Method | SOLUTION NMR / na | ||||||
 Authors Authors | Lee, Y. / Kim, Y. | ||||||
| Funding support |  Korea, Republic Of, 1items Korea, Republic Of, 1items
| ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2018 Journal: Biochemistry / Year: 2018Title: Tyr51: Key Determinant of the Low Thermostability of the Colwellia psychrerythraea Cold-Shock Protein. Authors: Lee, Y. / Kwak, C. / Jeong, K.W. / Durai, P. / Ryu, K.S. / Kim, E.H. / Cheong, C. / Ahn, H.C. / Kim, H.J. / Kim, Y. #1:  Journal: Biochem. Biophys. Res. Commun. / Year: 2014 Journal: Biochem. Biophys. Res. Commun. / Year: 2014Title: Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus Authors: Jin, B. / Jeong, K.W. / Kim, Y. #2:  Journal: Biochemistry / Year: 2013 Journal: Biochemistry / Year: 2013Title: Structural and dynamic features of cold-shock proteins of Listeria monocytogenes, a psychrophilic bacterium Authors: Lee, J. / Jeong, K.W. / Jin, B. / Ryu, K.S. / Kim, E.H. / Ahn, J.H. / Kim, Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
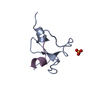
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5xv9.cif.gz 5xv9.cif.gz | 390.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5xv9.ent.gz pdb5xv9.ent.gz | 326.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5xv9.json.gz 5xv9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xv/5xv9 https://data.pdbj.org/pub/pdb/validation_reports/xv/5xv9 ftp://data.pdbj.org/pub/pdb/validation_reports/xv/5xv9 ftp://data.pdbj.org/pub/pdb/validation_reports/xv/5xv9 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7299.082 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Colwellia psychrerythraea (strain 34H / ATCC BAA-681) (bacteria) Colwellia psychrerythraea (strain 34H / ATCC BAA-681) (bacteria)Strain: 34H / ATCC BAA-681 / Gene: CPS_0718 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj HSQC
HSQC