[English] 日本語
 Yorodumi
Yorodumi- PDB-5ukz: NMR Solution structure of chemically synthesized antilisterial Pe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5ukz | ||||||
|---|---|---|---|---|---|---|---|
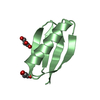
| Title | NMR Solution structure of chemically synthesized antilisterial Pediocin PA-1 M31L analog. | ||||||
 Components Components | Bacteriocin pediocin PA-1 M31L | ||||||
 Keywords Keywords | ANTIMICROBIAL PROTEIN / Antilisterial / Pediocin PA-1 / Pediococcus acidilactici UL5 / Bacteriocin / Chemical synthesis | ||||||
| Function / homology |  Function and homology information Function and homology informationkilling of cells of another organism / defense response to bacterium / extracellular region Similarity search - Function | ||||||
| Biological species |  Pediococcus acidilactici (bacteria) Pediococcus acidilactici (bacteria) | ||||||
| Method | SOLUTION NMR / na | ||||||
 Authors Authors | Bedard, F. / Hammami, R. / Zirah, S. / Rebuffat, S. / Fliss, I. / Biron, E. | ||||||
| Funding support |  Canada, 1items Canada, 1items
| ||||||
 Citation Citation |  Journal: Sci Rep / Year: 2018 Journal: Sci Rep / Year: 2018Title: Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof. Authors: Bedard, F. / Hammami, R. / Zirah, S. / Rebuffat, S. / Fliss, I. / Biron, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5ukz.cif.gz 5ukz.cif.gz | 128 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5ukz.ent.gz pdb5ukz.ent.gz | 103.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5ukz.json.gz 5ukz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uk/5ukz https://data.pdbj.org/pub/pdb/validation_reports/uk/5ukz ftp://data.pdbj.org/pub/pdb/validation_reports/uk/5ukz ftp://data.pdbj.org/pub/pdb/validation_reports/uk/5ukz | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4617.172 Da / Num. of mol.: 1 / Fragment: UNP residues 19-62 / Mutation: M31L / Source method: obtained synthetically / Details: Pediocin PA-1 producer / Source: (synth.)  Pediococcus acidilactici (bacteria) / References: UniProt: P29430 Pediococcus acidilactici (bacteria) / References: UniProt: P29430 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 3.33 mg/mL NA-1H Pediocin PA-1 M31L, trifluoroethanol/water Details: 300 uL of 0.1% TFA water plus 300 uL of 98% TFE-d2. Label: 1H_M31L / Solvent system: trifluoroethanol/water |
|---|---|
| Sample | Conc.: 3.33 mg/mL / Component: Pediocin PA-1 M31L / Isotopic labeling: NA-1H |
| Sample conditions | Details: 0.1% TFA was required to prevet disulfide bond rearrangment while acquisitions. TOCSY and NOESY spectra were recorded with mixing times of 80 ms and 300 ms and 16 and 72 scans, respectively. Ionic strength: 0 Not defined / Label: Conditions_1 / pH: 2.8 / PH err: 0.2 / Pressure: 1 atm / Temperature: 313 K / Temperature err: 0.1 |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE III / Manufacturer: Bruker / Model: AVANCE III / Field strength: 600 MHz / Details: Cyroprobe |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: na / Software ordinal: 2 Details: For this entry, the authors only used chemical shift values without constraints. The authors used the CS-ROSETTA protocol which allows structure determination of proteins based on chemical ...Details: For this entry, the authors only used chemical shift values without constraints. The authors used the CS-ROSETTA protocol which allows structure determination of proteins based on chemical shift information alone. | ||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 40000 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller





 PDBj
PDBj