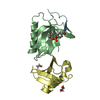
| Entry | Database: PDB / ID: 5mf2
|
|---|
| Title | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 |
|---|
 Components Components | Distal tail protein |
|---|
 Keywords Keywords | VIRAL PROTEIN / Lanthanide complexes / Crystallogenesis / Anomalous scattering / Bacteriophages |
|---|
| Function / homology |  Function and homology information Function and homology information
Elongation Factor Tu (Ef-tu); domain 3 - #290 / Phosphorylase Kinase; domain 1 - #190 / : / : / : / Distal tail protein pb9, A domain C-terminal / Distal tail protein pb9, A domain, N-terminal / Distal tail protein pb9, B domain / Elongation Factor Tu (Ef-tu); domain 3 / Phosphorylase Kinase; domain 1 ...Elongation Factor Tu (Ef-tu); domain 3 - #290 / Phosphorylase Kinase; domain 1 - #190 / : / : / : / Distal tail protein pb9, A domain C-terminal / Distal tail protein pb9, A domain, N-terminal / Distal tail protein pb9, B domain / Elongation Factor Tu (Ef-tu); domain 3 / Phosphorylase Kinase; domain 1 / Beta Barrel / 2-Layer Sandwich / Mainly Beta / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2 Å SYNCHROTRON / Resolution: 2 Å |
|---|
 Authors Authors | Engilberge, S. / Riobe, F. / Di Pietro, S. / Lassalle, L. / Arnaud, C.-A. / Breyton, C. / Madern, D. / Coquelle, N. / Maury, O. / Girard, E. |
|---|
| Funding support |  France, 1items France, 1items | Organization | Grant number | Country |
|---|
| French National Research Agency | ANR-13-BS07-0007-01 |  France France |
|
|---|
 Citation Citation |  Journal: Chem Sci / Year: 2017Title Journal: Chem Sci / Year: 2017Title: Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence. Authors: Engilberge, S. / Riobe, F. / Di Pietro, S. / Lassalle, L. / Coquelle, N. / Arnaud, C.A. / Pitrat, D. / Mulatier, J.C. / Madern, D. / Breyton, C. / Maury, O. / Girard, E. |
|---|
| History | | Deposition | Nov 17, 2016 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Sep 20, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Apr 18, 2018 | Group: Data collection / Database references / Category: citation / citation_author
Item: _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed ..._citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.name |
|---|
| Revision 2.0 | Jul 25, 2018 | Group: Atomic model / Data collection / Derived calculations
Category: atom_site / pdbx_struct_conn_angle
Item: _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ..._atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.value |
|---|
| Revision 2.1 | May 29, 2019 | Group: Data collection / Derived calculations / Structure summary
Category: chem_comp / entity / pdbx_entity_nonpoly
Item: _chem_comp.name / _entity.pdbx_description / _pdbx_entity_nonpoly.name |
|---|
| Revision 2.2 | Oct 16, 2019 | Group: Data collection / Category: reflns_shell |
|---|
| Revision 2.3 | May 8, 2024 | Group: Data collection / Database references / Derived calculations
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_struct_conn_angle / struct_conn
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_alt_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_comp_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_alt_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_ptnr1_label_alt_id / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_symmetry |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Escherichia phage T5 (virus)
Escherichia phage T5 (virus) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2 Å
SYNCHROTRON / Resolution: 2 Å  Authors
Authors France, 1items
France, 1items  Citation
Citation Journal: Chem Sci / Year: 2017
Journal: Chem Sci / Year: 2017 Journal: J. Virol. / Year: 2014
Journal: J. Virol. / Year: 2014 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5mf2.cif.gz
5mf2.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5mf2.ent.gz
pdb5mf2.ent.gz PDB format
PDB format 5mf2.json.gz
5mf2.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/mf/5mf2
https://data.pdbj.org/pub/pdb/validation_reports/mf/5mf2 ftp://data.pdbj.org/pub/pdb/validation_reports/mf/5mf2
ftp://data.pdbj.org/pub/pdb/validation_reports/mf/5mf2 Links
Links Assembly
Assembly




 Components
Components Escherichia phage T5 (virus) / Gene: D16, ORF128, T5.139, T5p135 / Production host:
Escherichia phage T5 (virus) / Gene: D16, ORF128, T5.139, T5p135 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID23-1 / Wavelength: 1.64 Å
/ Beamline: ID23-1 / Wavelength: 1.64 Å Processing
Processing Movie
Movie Controller
Controller











 PDBj
PDBj





