[English] 日本語
 Yorodumi
Yorodumi- PDB-5cbi: Crystal structure of Mycobacterium tuberculosis malate synthase i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5cbi | ||||||
|---|---|---|---|---|---|---|---|
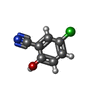
| Title | Crystal structure of Mycobacterium tuberculosis malate synthase in complex with 5-chloro-2-hydroxybenzonitrile | ||||||
 Components Components | Malate synthase G | ||||||
 Keywords Keywords | TRANSFERASE / Fragment / Complex / Acetyltransferase | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell extracellular matrix binding / capsule / malate synthase / malate synthase activity / glyoxylate catabolic process / coenzyme A binding / adhesion of symbiont to host / coenzyme A metabolic process / glyoxylate cycle / fibronectin binding ...host cell extracellular matrix binding / capsule / malate synthase / malate synthase activity / glyoxylate catabolic process / coenzyme A binding / adhesion of symbiont to host / coenzyme A metabolic process / glyoxylate cycle / fibronectin binding / tricarboxylic acid cycle / laminin binding / peptidoglycan-based cell wall / cell surface / magnesium ion binding / protein homodimerization activity / extracellular region / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.988 Å MOLECULAR REPLACEMENT / Resolution: 1.988 Å | ||||||
 Authors Authors | Huang, H.-L. / Sacchettini, J.C. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2016 Journal: J. Biol. Chem. / Year: 2016Title: Mycobacterium tuberculosis Malate Synthase Structures with Fragments Reveal a Portal for Substrate/Product Exchange. Authors: Huang, H.L. / Krieger, I.V. / Parai, M.K. / Gawandi, V.B. / Sacchettini, J.C. #1:  Journal: J.Biol.Chem. / Year: 2003 Journal: J.Biol.Chem. / Year: 2003Title: Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. Authors: Smith, C.V. / Huang, C.C. / Miczak, A. / Russell, D.G. / Sacchettini, J.C. / Honer zu Bentrup, K. #2:  Journal: Chem.Biol. / Year: 2012 Journal: Chem.Biol. / Year: 2012Title: Structure-guided discovery of phenyl-diketo acids as potent inhibitors of M. tuberculosis malate synthase. Authors: Krieger, I.V. / Freundlich, J.S. / Gawandi, V.B. / Roberts, J.P. / Gawandi, V.B. / Sun, Q. / Owen, J.L. / Fraile, M.T. / Huss, S.I. / Lavandera, J.L. / Ioerger, T.R. / Sacchettini, J.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5cbi.cif.gz 5cbi.cif.gz | 163 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5cbi.ent.gz pdb5cbi.ent.gz | 125.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5cbi.json.gz 5cbi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cb/5cbi https://data.pdbj.org/pub/pdb/validation_reports/cb/5cbi ftp://data.pdbj.org/pub/pdb/validation_reports/cb/5cbi ftp://data.pdbj.org/pub/pdb/validation_reports/cb/5cbi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5c7vC  5c9rC  5c9uC  5c9wC  5c9xC  5cahC  5cakC  5cbbC  5cbjC  5cczC  5cewC  5cjmC  5cjnC  5drcC  5driC  5dx7C  5e9xC  5ecvC  5h8mC  5h8pC  5h8uC  5t8gC  1n8iS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 80456.734 Da / Num. of mol.: 1 / Mutation: C619A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv / Gene: glcB, Rv1837c, MTCY1A11.06 / Production host:  | ||
|---|---|---|---|
| #2: Chemical | ChemComp-MG / | ||
| #3: Chemical | ChemComp-4ZC / #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 44.76 % |
|---|---|
| Crystal grow | Temperature: 300 K / Method: vapor diffusion, hanging drop / pH: 7.5 / Details: PEG 3350, magnesium chloride, tris / PH range: 7.0-8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97924 Å / Beamline: 19-ID / Wavelength: 0.97924 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Dec 11, 2009 |
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97924 Å / Relative weight: 1 |
| Reflection | Resolution: 1.988→46.09 Å / Num. all: 51080 / Num. obs: 50244 / % possible obs: 98.4 % / Redundancy: 8.7 % / Biso Wilson estimate: 35.349 Å2 / Rsym value: 0.028 / Net I/σ(I): 18.66 |
| Reflection shell | Resolution: 1.988→2.1 Å / Redundancy: 4.7 % / % possible all: 92 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1N8I Resolution: 1.988→45.096 Å / SU ML: 0.23 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 27.72 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.988→45.096 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller



















 PDBj
PDBj