[English] 日本語
 Yorodumi
Yorodumi- PDB-5adk: Structure of bovine endothelial nitric oxide synthase heme domain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5adk | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((3-(Dimethylamino)methyl)phenoxy)methyl)quinolin-2- amine | ||||||
 Components Components | NITRIC OXIDE SYNTHASE, ENDOTHELIAL | ||||||
 Keywords Keywords | OXIDOREDUCTASE / NITRIC OXIDE SYNTHASE / INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of relaxation of smooth muscle / cellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone ...positive regulation of relaxation of smooth muscle / cellular response to laminar fluid shear stress / negative regulation of leukocyte cell-cell adhesion / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / nitric-oxide synthase activity / L-arginine catabolic process / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of blood pressure / response to hormone / nitric oxide biosynthetic process / mitochondrion organization / caveola / response to peptide hormone / blood coagulation / FMN binding / NADP binding / flavin adenine dinucleotide binding / response to lipopolysaccharide / cytoskeleton / calmodulin binding / heme binding / Golgi apparatus / metal ion binding / nucleus / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.805 Å MOLECULAR REPLACEMENT / Resolution: 1.805 Å | ||||||
 Authors Authors | Li, H. / Poulos, T.L. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2015 Journal: J.Med.Chem. / Year: 2015Title: Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase. Authors: Cinelli, M.A. / Li, H. / Pensa, A.V. / Kang, S. / Roman, L.J. / Martasek, P. / Poulos, T.L. / Silverman, R.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5adk.cif.gz 5adk.cif.gz | 354.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5adk.ent.gz pdb5adk.ent.gz | 286.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5adk.json.gz 5adk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ad/5adk https://data.pdbj.org/pub/pdb/validation_reports/ad/5adk ftp://data.pdbj.org/pub/pdb/validation_reports/ad/5adk ftp://data.pdbj.org/pub/pdb/validation_reports/ad/5adk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5ad4C  5ad5C  5ad6C  5ad7C  5ad8C  5ad9C  5adaC  5adbC  5adcC  5addC  5adeC  5adfC  5adgC  5adiC  5adjC  5adlC  5admC  5adnC  5fj2C  5fj3C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 49727.012 Da / Num. of mol.: 2 / Fragment: HEME DOMAIN, UNP RESIDUES 40-482 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|
-Non-polymers , 7 types, 515 molecules 

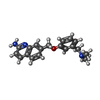










| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-ACT / #6: Chemical | #7: Chemical | ChemComp-ZN / | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | RESIDUE 100 IS FOUND AS AN ARG IN STRUCTURE BUT IS A CYS IN DATABASE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.45 Å3/Da / Density % sol: 49.8 % Description: OVERALL RMERGE 0.082 RPIM0.056 CC ONE HALF 0.999 HIGHEST RESOLUTION SHELL RMERGE 3.157 RPIM 2.226 CC ONE HALF 0.260 |
|---|---|
| Crystal grow | pH: 6 Details: 20-22% PEG3350 0.1M CACODYLATE, 140-200 MM MG ACETATE 5 MM TCEP, pH 6.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL12-2 / Wavelength: 0.979 / Beamline: BL12-2 / Wavelength: 0.979 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 31, 2014 / Details: MIRRORS |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 1.81→50 Å / Num. obs: 89217 / % possible obs: 99.2 % / Observed criterion σ(I): -3 / Redundancy: 5.5 % / Biso Wilson estimate: 32.45 Å2 / Rmerge(I) obs: 0.08 / Net I/σ(I): 12 |
| Reflection shell | Resolution: 1.81→1.85 Å / Redundancy: 4.9 % / Rmerge(I) obs: 1.5 / Mean I/σ(I) obs: 0.5 / % possible all: 95.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.805→39.19 Å / SU ML: 0.23 / σ(F): 1.33 / Phase error: 20.82 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 1.805→39.19 Å / SU ML: 0.23 / σ(F): 1.33 / Phase error: 20.82 / Stereochemistry target values: MLDetails: RESIDUES 110 TO 120 IN CHAIN A AND 111 TO 120 IN CHAIN B ARE DISORDERED.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.805→39.19 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj








