[English] 日本語
 Yorodumi
Yorodumi- PDB-4rsk: STRUCTURE OF THE K7A/R10A/K66A VARIANT OF RIBONUCLEASE A COMPLEXE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4rsk | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF THE K7A/R10A/K66A VARIANT OF RIBONUCLEASE A COMPLEXED WITH 3'-UMP | ||||||
 Components Components | RIBONUCLEASE A | ||||||
 Keywords Keywords | HYDROLASE / ENDONUCLEASE / RIBONUCLEASE A / SITE-DIRECTED MUTAGENESIS | ||||||
| Function / homology |  Function and homology information Function and homology informationpancreatic ribonuclease / ribonuclease A activity / RNA nuclease activity / nucleic acid binding / defense response to Gram-positive bacterium / lyase activity / extracellular region Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / DIFFERENCE FOURIER / Resolution: 2.1 Å X-RAY DIFFRACTION / DIFFERENCE FOURIER / Resolution: 2.1 Å | ||||||
 Authors Authors | Schultz, L.W. / Fisher, B.M. / Raines, R.T. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Coulombic Effects of Remote Subsites on the Active Site of Ribonuclease A Authors: Fisher, B.M. / Schultz, L.W. / Raines, R.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
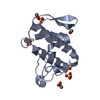
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4rsk.cif.gz 4rsk.cif.gz | 39.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4rsk.ent.gz pdb4rsk.ent.gz | 26.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4rsk.json.gz 4rsk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rs/4rsk https://data.pdbj.org/pub/pdb/validation_reports/rs/4rsk ftp://data.pdbj.org/pub/pdb/validation_reports/rs/4rsk ftp://data.pdbj.org/pub/pdb/validation_reports/rs/4rsk | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3rskSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 13506.006 Da / Num. of mol.: 1 / Mutation: K7A, R10A, K66A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Chemical | ChemComp-U3P / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.47 Å3/Da / Density % sol: 60 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 4.5 Details: CRYSTALS WERE PREPARED USING THE HANGING DROP METHOD. LYOPHILIZED K7A/R10A/K66A RNASE A WAS DISSOLVED IN UNBUFFERED WATER TO A CONCENTRATION OF 60MG/ML. DROPS CONSISTING OF PROTEIN SOLUTION ...Details: CRYSTALS WERE PREPARED USING THE HANGING DROP METHOD. LYOPHILIZED K7A/R10A/K66A RNASE A WAS DISSOLVED IN UNBUFFERED WATER TO A CONCENTRATION OF 60MG/ML. DROPS CONSISTING OF PROTEIN SOLUTION (1.5MICROLITER), WATER (1.5MICROLITER), AND RESERVOIR SOLUTION (3.0MICROLITER) WERE SUSPENDED OVER 0.5 ML OF RESERVOIR SOLUTION (0.1M SODIUM ACETATE BUFFER, PH 4.5, CONTAINING 36% PEG 4000). THE COMPLEX WITH 3'-UMP WAS PREPARED BY SOAKING CRYSTALS IN MOTHER LIQUOR CONTAINING 5MM 3'-UMP FOR 2 DAYS., vapor diffusion - hanging drop | ||||||||||||||||||||||||||||||
| Crystal | *PLUS | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 273 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Sep 12, 1997 / Details: LONG MIRRORS |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→30 Å / Num. obs: 33552 / % possible obs: 88 % / Observed criterion σ(I): 0.33 / Redundancy: 2.4 % / Rsym value: 0.026 / Net I/σ(I): 19.8 |
| Reflection shell | Resolution: 2.1→2.2 Å / Rsym value: 0.12 / % possible all: 88 |
| Reflection | *PLUS Num. obs: 14210 / Num. measured all: 33552 / Rmerge(I) obs: 0.026 |
| Reflection shell | *PLUS % possible obs: 88 % / Rmerge(I) obs: 0.122 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: DIFFERENCE FOURIER Starting model: PDB ENTRY 3RSK Resolution: 2.1→30 Å / Isotropic thermal model: TNT BCORREL / σ(F): 0 / Stereochemistry target values: TNT PROTGEO
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: BABINET SCALING / Bsol: 135 Å2 / ksol: 0.56 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Version: 5E / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj