[English] 日本語
 Yorodumi
Yorodumi- PDB-4l70: Human artd3 (parp3) - catalytic domain in complex with inhibitor ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4l70 | ||||||
|---|---|---|---|---|---|---|---|
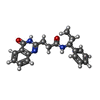
| Title | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0352 | ||||||
 Components Components | Poly [ADP-ribose] polymerase 3 | ||||||
 Keywords Keywords | transferase/transferase inhibitor / DIPHTHERIA TOXIN LIKE ADP-RIBOSE TRANSFERASE / TRANSFERASE / ADP-RIBOSYLATION / transferase-transferase inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of isotype switching / NAD+-protein-lysine ADP-ribosyltransferase activity / NAD DNA ADP-ribosyltransferase activity / DNA ADP-ribosylation / protein localization to site of double-strand break / protein auto-ADP-ribosylation / NAD+-protein-aspartate ADP-ribosyltransferase activity / positive regulation of double-strand break repair via nonhomologous end joining / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity ...negative regulation of isotype switching / NAD+-protein-lysine ADP-ribosyltransferase activity / NAD DNA ADP-ribosyltransferase activity / DNA ADP-ribosylation / protein localization to site of double-strand break / protein auto-ADP-ribosylation / NAD+-protein-aspartate ADP-ribosyltransferase activity / positive regulation of double-strand break repair via nonhomologous end joining / NAD+-protein-glutamate ADP-ribosyltransferase activity / NAD+-protein mono-ADP-ribosyltransferase activity / negative regulation of telomere maintenance via telomerase / Transferases; Glycosyltransferases; Pentosyltransferases / catalytic activity / NAD+ poly-ADP-ribosyltransferase activity / intercellular bridge / nucleotidyltransferase activity / centriole / telomere maintenance / regulation of mitotic spindle organization / double-strand break repair via nonhomologous end joining / double-strand break repair / site of double-strand break / nuclear body / centrosome / nucleolus / nucleoplasm / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Karlberg, T. / Thorsell, A.G. / Lindgren, A.E.G. / Ekblad, T. / Spjut, S. / Andersson, C.D. / Weigelt, J. / Linusson, A. / Elofsson, M. / Schuler, H. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2013 Journal: J.Med.Chem. / Year: 2013Title: Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3. Authors: Lindgren, A.E. / Karlberg, T. / Ekblad, T. / Spjut, S. / Thorsell, A.G. / Andersson, C.D. / Nhan, T.T. / Hellsten, V. / Weigelt, J. / Linusson, A. / Schuler, H. / Elofsson, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4l70.cif.gz 4l70.cif.gz | 148.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4l70.ent.gz pdb4l70.ent.gz | 115.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4l70.json.gz 4l70.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l7/4l70 https://data.pdbj.org/pub/pdb/validation_reports/l7/4l70 ftp://data.pdbj.org/pub/pdb/validation_reports/l7/4l70 ftp://data.pdbj.org/pub/pdb/validation_reports/l7/4l70 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4l6zC  4l7lC  4l7nC  4l7oC  4l7pC  4l7rC  4l7uC  4gv4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 39752.074 Da / Num. of mol.: 1 / Fragment: CATALYTIC PARP DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ADPRT3, ADPRTL3, PARP3 / Plasmid: pNIC-Bsa4 / Production host: Homo sapiens (human) / Gene: ADPRT3, ADPRTL3, PARP3 / Plasmid: pNIC-Bsa4 / Production host:  | ||
|---|---|---|---|
| #2: Chemical | ChemComp-1V9 / | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.56 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1.6M DL-Malic Acid, 0.1M Bis-tris-propane , pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9919 Å / Beamline: I04 / Wavelength: 0.9919 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 27, 2011 |
| Radiation | Monochromator: double crystal monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9919 Å / Relative weight: 1 |
| Reflection | Resolution: 2→30 Å / Num. all: 22105 / Num. obs: 22105 / % possible obs: 99.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 7.4 % / Rmerge(I) obs: 0.13 / Rsym value: 0.13 / Net I/σ(I): 15.6 |
| Reflection shell | Resolution: 2→2.05 Å / Redundancy: 7.2 % / Rmerge(I) obs: 0.79 / Mean I/σ(I) obs: 3.1 / Num. unique all: 1634 / Rsym value: 0.58 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4GV4 Resolution: 2→28.5 Å / Cor.coef. Fo:Fc: 0.955 / Cor.coef. Fo:Fc free: 0.935 / SU B: 8.3 / SU ML: 0.107 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.19 / ESU R Free: 0.165 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.944 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→28.5 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2→2.052 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 18.54 Å / Origin y: 0.575 Å / Origin z: 11.902 Å
|
 Movie
Movie Controller
Controller













 PDBj
PDBj