[English] 日本語
 Yorodumi
Yorodumi- PDB-4l6y: Structure of the microtubule associated protein PRC1 (Protein Reg... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4l6y | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of the microtubule associated protein PRC1 (Protein Regulator of Cytokinesis 1) | ||||||
 Components Components | Protein regulator of cytokinesis 1 | ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Spectrin / Microtubule binding / microtubule crosslinking / microtubule / kinesin-4 / spindle midzone | ||||||
| Function / homology |  Function and homology information Function and homology informationcontractile ring / mitotic spindle midzone / mitotic spindle elongation / mitotic spindle midzone assembly / RHO GTPases activate CIT / kinesin binding / intercellular bridge / regulation of cytokinesis / spindle microtubule / microtubule cytoskeleton organization ...contractile ring / mitotic spindle midzone / mitotic spindle elongation / mitotic spindle midzone assembly / RHO GTPases activate CIT / kinesin binding / intercellular bridge / regulation of cytokinesis / spindle microtubule / microtubule cytoskeleton organization / spindle / spindle pole / chromosome / microtubule cytoskeleton / midbody / microtubule binding / cell division / positive regulation of cell population proliferation / protein kinase binding / nucleoplasm / identical protein binding / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.3015 Å MOLECULAR REPLACEMENT / Resolution: 3.3015 Å | ||||||
 Authors Authors | Subramanian, R. / Ti, S. / Tan, L. / Darst, S.A. / Kapoor, T.M. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 2013 Journal: Cell(Cambridge,Mass.) / Year: 2013Title: Marking and Measuring Single Microtubules by PRC1 and Kinesin-4. Authors: Subramanian, R. / Ti, S.C. / Tan, L. / Darst, S.A. / Kapoor, T.M. | ||||||
| History |
| ||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4l6y.cif.gz 4l6y.cif.gz | 178.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4l6y.ent.gz pdb4l6y.ent.gz | 138.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4l6y.json.gz 4l6y.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4l6y_validation.pdf.gz 4l6y_validation.pdf.gz | 430.6 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4l6y_full_validation.pdf.gz 4l6y_full_validation.pdf.gz | 436.1 KB | Display | |
| Data in XML |  4l6y_validation.xml.gz 4l6y_validation.xml.gz | 29.2 KB | Display | |
| Data in CIF |  4l6y_validation.cif.gz 4l6y_validation.cif.gz | 39.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l6/4l6y https://data.pdbj.org/pub/pdb/validation_reports/l6/4l6y ftp://data.pdbj.org/pub/pdb/validation_reports/l6/4l6y ftp://data.pdbj.org/pub/pdb/validation_reports/l6/4l6y | HTTPS FTP |
-Related structure data
| Related structure data |  4l3iSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 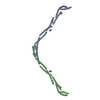
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 58182.660 Da / Num. of mol.: 2 / Fragment: UNP Residues 1-486 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PRC1 / Production host: Homo sapiens (human) / Gene: PRC1 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 6.3 Å3/Da / Density % sol: 80.5 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, hanging drop / pH: 8 Details: Concentration: 5-10 mg/ml. Protein buffer: 80 mM PIPES (pH 6.8), 1 mM MgCl2, 1 mM EGTA, 150 mM KCl, VAPOR DIFFUSION, HANGING DROP, temperature 294K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-E / Wavelength: 0.9792 Å / Beamline: 24-ID-E / Wavelength: 0.9792 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jul 1, 2011 |
| Radiation | Monochromator: Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9792 Å / Relative weight: 1 |
| Reflection twin | Operator: h,-k,-h-l / Fraction: 0.5 |
| Reflection | Resolution: 3.2→40 Å / Num. all: 40830 / Num. obs: 39522 / % possible obs: 90.5 % / Observed criterion σ(F): 1.4 / Observed criterion σ(I): 1.4 |
| Reflection shell | Resolution: 3.2→3.31 Å / % possible all: 82.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4L3I Resolution: 3.3015→29.906 Å / σ(F): 1.37 / Phase error: 35.46 / Stereochemistry target values: TWIN_LSQ_F
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.3015→29.906 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj


