[English] 日本語
 Yorodumi
Yorodumi- PDB-4l5k: Crystal structure of the complex of DNA hexamer d(CGATCG) with Co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4l5k | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
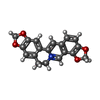
| Title | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / DRUG-DNA COMPLEX / Isoquinoline alkaloid | Function / homology | Chem-KPT / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.71 Å molecular replacement / Resolution: 2.71 Å  Authors AuthorsFerraroni, M. / Bazzicalupi, C. / Gratteri, P. |  Citation Citation Journal: to be published Journal: to be publishedTitle: Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine Authors: Ferraroni, M. / Bazzicalupi, C. / Gratteri, P. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4l5k.cif.gz 4l5k.cif.gz | 14 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4l5k.ent.gz pdb4l5k.ent.gz | 7.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4l5k.json.gz 4l5k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l5/4l5k https://data.pdbj.org/pub/pdb/validation_reports/l5/4l5k ftp://data.pdbj.org/pub/pdb/validation_reports/l5/4l5k ftp://data.pdbj.org/pub/pdb/validation_reports/l5/4l5k | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3ft6S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: DNA chain | Mass: 1809.217 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: Chemical | ChemComp-KPT / |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43.45 % |
|---|---|
| Crystal grow | Temperature: 296 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 10% MPD, 40 mM Na Cacodylate, 12 mM spermine tetra-HCl, 80 mM sodium chloride and 20 mM barium chloride, pH 6.0, vapor diffusion, sitting drop, temperature 296K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source: SEALED TUBE / Type: OXFORD DIFFRACTION ENHANCE ULTRA / Wavelength: 1.542 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: OXFORD ONYX CCD / Detector: CCD / Date: Aug 14, 2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.542 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.7→14.78 Å / Num. obs: 558 / % possible obs: 97 % / Observed criterion σ(I): -3 / Rmerge(I) obs: 0.092 / Net I/σ(I): 18.99 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 3FT6 Resolution: 2.71→14.78 Å / Cor.coef. Fo:Fc: 0.932 / Occupancy max: 1 / Occupancy min: 0.5 / SU B: 28.317 / SU ML: 0.447 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 93.273 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.71→14.78 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj