| Entry | Database: PDB / ID: 4l48
|
|---|
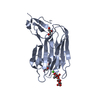
| Title | Crystal structure of d78n mutant clavibacter michiganensis expansin in complex with cellohexaose |
|---|
 Components Components | cellulose binding protein |
|---|
 Keywords Keywords | SUGAR BINDING PROTEIN / Cellulose binding protein / Cell wall loosening |
|---|
| Function / homology |  Function and homology information Function and homology information
: / Expansin, cellulose-binding-like domain / Expansin C-terminal domain / Expansin, cellulose-binding-like domain / RlpA-like protein, double-psi beta-barrel domain / Lytic transglycolase / Expansin, cellulose-binding-like domain superfamily / RlpA-like domain / RlpA-like domain superfamily / Cellulose binding domain ...: / Expansin, cellulose-binding-like domain / Expansin C-terminal domain / Expansin, cellulose-binding-like domain / RlpA-like protein, double-psi beta-barrel domain / Lytic transglycolase / Expansin, cellulose-binding-like domain superfamily / RlpA-like domain / RlpA-like domain superfamily / Cellulose binding domain / Carbohydrate-binding type-2 domain / CBM2 (Carbohydrate-binding type-2) domain profile. / CBD_II / CBM2, carbohydrate-binding domain superfamily / Glycoside hydrolase, family 5, conserved site / Glycosyl hydrolases family 5 signature. / CBM2/CBM3, carbohydrate-binding domain superfamily / Barwin-like endoglucanases / Cellulase (glycosyl hydrolase family 5) / Glycoside hydrolase, family 5 / Glycoside hydrolase superfamily / Beta Barrel / Immunoglobulin-like / Sandwich / Mainly BetaSimilarity search - Domain/homology |
|---|
| Biological species |  Clavibacter michiganensis subsp. michiganensis (bacteria) Clavibacter michiganensis subsp. michiganensis (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å |
|---|
 Authors Authors | Yennawar, N.H. / Yennawar, H.P. / Georgelis, N. / Cosgrove, D. |
|---|
 Citation Citation |  Journal: To be Published Journal: To be Published
Title: Crystal structure of wild type and d78n mutant clavibacter michiganensis expansin, in apo form and in complex with oligosaccharides
Authors: Yennawar, N.H. / Yennawar, H.P. / Georgelis, N. / Cosgrove, D. |
|---|
| History | | Deposition | Jun 7, 2013 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Mar 26, 2014 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jun 24, 2015 | Group: Structure summary |
|---|
| Revision 1.2 | Oct 21, 2015 | Group: Structure summary |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Data collection ...Atomic model / Data collection / Database references / Derived calculations / Non-polymer description / Structure summary
Category: atom_site / chem_comp ...atom_site / chem_comp / entity / entity_name_com / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_molecule_features / pdbx_nonpoly_scheme / struct_conn / struct_conn_type / struct_ref_seq_dif / struct_site / struct_site_gen
Item: _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ..._atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.auth_seq_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.type_symbol / _chem_comp.formula / _chem_comp.formula_weight / _chem_comp.id / _chem_comp.name / _chem_comp.type / _entity.formula_weight / _entity.pdbx_description / _entity.type / _struct_ref_seq_dif.details
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Sep 20, 2023 | Group: Data collection / Database references ...Data collection / Database references / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 2.2 | Nov 6, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Clavibacter michiganensis subsp. michiganensis (bacteria)
Clavibacter michiganensis subsp. michiganensis (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å
MOLECULAR REPLACEMENT / Resolution: 2.1 Å  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4l48.cif.gz
4l48.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4l48.ent.gz
pdb4l48.ent.gz PDB format
PDB format 4l48.json.gz
4l48.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/l4/4l48
https://data.pdbj.org/pub/pdb/validation_reports/l4/4l48 ftp://data.pdbj.org/pub/pdb/validation_reports/l4/4l48
ftp://data.pdbj.org/pub/pdb/validation_reports/l4/4l48
 Links
Links Assembly
Assembly


 Components
Components Clavibacter michiganensis subsp. michiganensis (bacteria)
Clavibacter michiganensis subsp. michiganensis (bacteria)
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller













 PDBj
PDBj


