[English] 日本語
 Yorodumi
Yorodumi- PDB-4bil: Threading model of the T7 large terminase within the gp8gp19 complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4bil | ||||||
|---|---|---|---|---|---|---|---|
| Title | Threading model of the T7 large terminase within the gp8gp19 complex | ||||||
 Components Components | DNA MATURASE B | ||||||
 Keywords Keywords | HYDROLASE / PACKAGING MOTOR / CONNECTOR / DNA TRANSLOCATION / ATPASE. | ||||||
| Function / homology |  Function and homology information Function and homology informationviral terminase, large subunit / viral DNA genome packaging / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / chromosome organization / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / endonuclease activity / ATP hydrolysis activity / ATP binding / metal ion binding Similarity search - Function | ||||||
| Biological species |   ENTEROBACTERIA PHAGE T7 (virus) ENTEROBACTERIA PHAGE T7 (virus) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / negative staining / Resolution: 29 Å | ||||||
 Authors Authors | Dauden, M.I. / Martin-Benito, J. / Sanchez-Ferrero, J.C. / Pulido-Cid, M. / Valpuesta, J.M. / Carrascosa, J.L. | ||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2013 Journal: J Biol Chem / Year: 2013Title: Large terminase conformational change induced by connector binding in bacteriophage T7. Authors: María I Daudén / Jaime Martín-Benito / Juan C Sánchez-Ferrero / Mar Pulido-Cid / José M Valpuesta / José L Carrascosa /  Abstract: During bacteriophage morphogenesis DNA is translocated into a preformed prohead by the complex formed by the portal protein, or connector, plus the terminase, which are located at an especial prohead ...During bacteriophage morphogenesis DNA is translocated into a preformed prohead by the complex formed by the portal protein, or connector, plus the terminase, which are located at an especial prohead vertex. The terminase is a powerful motor that converts ATP hydrolysis into mechanical movement of the DNA. Here, we have determined the structure of the T7 large terminase by electron microscopy. The five terminase subunits assemble in a toroid that encloses a channel wide enough to accommodate dsDNA. The structure of the complete connector-terminase complex is also reported, revealing the coupling between the terminase and the connector forming a continuous channel. The structure of the terminase assembled into the complex showed a different conformation when compared with the isolated terminase pentamer. To understand in molecular terms the terminase morphological change, we generated the terminase atomic model based on the crystallographic structure of its phage T4 counterpart. The docking of the threaded model in both terminase conformations showed that the transition between the two states can be achieved by rigid body subunit rotation in the pentameric assembly. The existence of two terminase conformations and its possible relation to the sequential DNA translocation may shed light into the molecular bases of the packaging mechanism of bacteriophage T7. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bil.cif.gz 4bil.cif.gz | 405.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bil.ent.gz pdb4bil.ent.gz | 323.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bil.json.gz 4bil.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4bil_validation.pdf.gz 4bil_validation.pdf.gz | 802.2 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4bil_full_validation.pdf.gz 4bil_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  4bil_validation.xml.gz 4bil_validation.xml.gz | 145.7 KB | Display | |
| Data in CIF |  4bil_validation.cif.gz 4bil_validation.cif.gz | 196.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bi/4bil https://data.pdbj.org/pub/pdb/validation_reports/bi/4bil ftp://data.pdbj.org/pub/pdb/validation_reports/bi/4bil ftp://data.pdbj.org/pub/pdb/validation_reports/bi/4bil | HTTPS FTP |
-Related structure data
| Related structure data |  2356MC 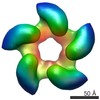 2355C  4bijC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 53822.504 Da / Num. of mol.: 5 / Fragment: RESIDUES 1-476 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   ENTEROBACTERIA PHAGE T7 (virus) / Production host: ENTEROBACTERIA PHAGE T7 (virus) / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: GP8GP19 COMPLEX OF BACTERIOPHAGE T7 / Type: VIRUS |
|---|---|
| Buffer solution | Name: 50 MM SODIUM PHOSPHATE BUFFER PH 7, 300 MM NACL, 10 MM MGCL2, 1 MM ADP 5 MM DTT AND 20% (V/V) GLYCEROL pH: 7.4 Details: 50 MM SODIUM PHOSPHATE BUFFER PH 7, 300 MM NACL, 10 MM MGCL2, 1 MM ADP 5 MM DTT AND 20% (V/V) GLYCEROL |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: YES / Vitrification applied: NO |
| EM staining | Type: NEGATIVE / Material: uranyl acetate |
| Specimen support | Details: CARBON |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TECNAI F20 / Date: Jul 7, 2011 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 67000 X / Nominal defocus max: 3200 nm / Nominal defocus min: 1500 nm / Cs: 2.26 mm |
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) |
| Image scans | Num. digital images: 573 |
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| EM software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: EACH PLATE | ||||||||||||||||||
| Symmetry | Point symmetry: C5 (5 fold cyclic) | ||||||||||||||||||
| 3D reconstruction | Method: COMMON LINES AND PROJECTION MATCHING / Resolution: 29 Å / Num. of particles: 837 / Actual pixel size: 4.2 Å Details: 3CPE PDB WAS USED AS TEMPLATE FOR THE GENERATION OF THE THREADING MODEL OF GP19 WITHIN THE GP19GP8 COMPLEX SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-2356. (DEPOSITION ID: 11615). Symmetry type: POINT | ||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL Details: METHOD--RIGID BODY REFINEMENT PROTOCOL--THREADING MODEL | ||||||||||||||||||
| Atomic model building | PDB-ID: 3CPE Accession code: 3CPE / Source name: PDB / Type: experimental model | ||||||||||||||||||
| Refinement | Highest resolution: 29 Å | ||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 29 Å
|
 Movie
Movie Controller
Controller



 PDBj
PDBj
