[English] 日本語
 Yorodumi
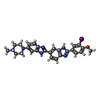
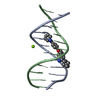
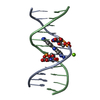
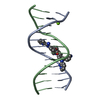
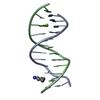
Yorodumi- PDB-442d: 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIV... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 442d | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / B-DNA / MINOR GROOVE BINDING | Function / homology | Chem-IB / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / Resolution: 1.6 Å X-RAY DIFFRACTION / Resolution: 1.6 Å  Authors AuthorsClark, G.R. / Squire, C.J. / Martin, R.F. / White, J. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2000 Journal: Nucleic Acids Res. / Year: 2000Title: Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design. Authors: Squire, C.J. / Baker, L.J. / Clark, G.R. / Martin, R.F. / White, J. #1:  Journal: Nucleic Acids Res. / Year: 2000 Journal: Nucleic Acids Res. / Year: 2000Title: Intermolecular interactions and water structure in a condensed phase B-DNA crystal Authors: Clark, G.R. / Squire, C.J. / Baker, L.J. / Martin, R.F. / White, J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  442d.cif.gz 442d.cif.gz | 28 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb442d.ent.gz pdb442d.ent.gz | 17.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  442d.json.gz 442d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/42/442d https://data.pdbj.org/pub/pdb/validation_reports/42/442d ftp://data.pdbj.org/pub/pdb/validation_reports/42/442d ftp://data.pdbj.org/pub/pdb/validation_reports/42/442d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  443dC  444dC  445dC  447dC  448dC  449dC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3663.392 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | ChemComp-IB / | #3: Chemical | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.73 Å3/Da / Density % sol: 29.09 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 Details: pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: SIEMENS / Wavelength: 0.7107 ROTATING ANODE / Type: SIEMENS / Wavelength: 0.7107 |
| Detector | Type: BRUKER SMART / Detector: CCD / Date: Sep 27, 1997 |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.7107 Å / Relative weight: 1 |
| Reflection | Highest resolution: 1.6 Å / Num. all: 10398 / Num. obs: 10398 / % possible obs: 97.5 % / Observed criterion σ(F): 0 / Redundancy: 5.04 % / Rmerge(I) obs: 0.076 |
| Reflection shell | Resolution: 1.6→1.657 Å / Rmerge(I) obs: 0.252 |
| Reflection | *PLUS Num. obs: 9015 / Observed criterion σ(F): 2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.6→8 Å / Cross valid method: THROUGHOUT / σ(F): 4
| |||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→8 Å
| |||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||
| Software | *PLUS Name: SHELXL-97 / Classification: refinement | |||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.167 | |||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller










 PDBj
PDBj