[English] 日本語
 Yorodumi
Yorodumi- PDB-3tci: Crystal structure of the decameric sequence d(CGGGCGCCCG) as Z ty... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3tci | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the decameric sequence d(CGGGCGCCCG) as Z type duplex | ||||||||||||||||||
 Components Components | (DNA (5'-D(P* Keywords KeywordsDNA / Z-DNA duplex | Function / homology | DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.421 Å MOLECULAR REPLACEMENT / Resolution: 2.421 Å  Authors AuthorsVenkadesh, S. / Mandal, P.K. / Gautham, N. |  Citation Citation Journal: To be Published Journal: To be PublishedTitle: Crystal structure of the decameric sequence d(CGGGCGCCCG) as Z type duplex Authors: Venkadesh, S. / Mandal, P.K. / Gautham, N. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3tci.cif.gz 3tci.cif.gz | 15.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3tci.ent.gz pdb3tci.ent.gz | 10.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3tci.json.gz 3tci.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3tci_validation.pdf.gz 3tci_validation.pdf.gz | 377.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3tci_full_validation.pdf.gz 3tci_full_validation.pdf.gz | 377.8 KB | Display | |
| Data in XML |  3tci_validation.xml.gz 3tci_validation.xml.gz | 2.8 KB | Display | |
| Data in CIF |  3tci_validation.cif.gz 3tci_validation.cif.gz | 3.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tc/3tci https://data.pdbj.org/pub/pdb/validation_reports/tc/3tci ftp://data.pdbj.org/pub/pdb/validation_reports/tc/3tci ftp://data.pdbj.org/pub/pdb/validation_reports/tc/3tci | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 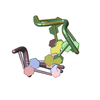
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 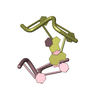
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 1191.818 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: Chemically synthesized #2: DNA chain | Mass: 1191.818 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: Chemically synthesized #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 1mM DNA, 2M MgCl2, 1mM spermine, 50% MPD , pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: BRUKER AXS MICROSTAR / Wavelength: 1.5418 Å ROTATING ANODE / Type: BRUKER AXS MICROSTAR / Wavelength: 1.5418 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Apr 2, 2009 / Details: mirrors |
| Radiation | Monochromator: mirrors / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.41→30 Å / Num. all: 586 / Num. obs: 579 / % possible obs: 98.8 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Redundancy: 3.41 % / Biso Wilson estimate: 54.506 Å2 / Rmerge(I) obs: 0.061 / Rsym value: 0.053 / Net I/σ(I): 4.4 |
| Reflection shell | Resolution: 2.41→2.5 Å / Redundancy: 3.51 % / Rmerge(I) obs: 0.271 / Mean I/σ(I) obs: 1.7 / Num. unique all: 63 / Rsym value: 0.224 / % possible all: 96.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.421→15.387 Å / SU ML: 0.33 / σ(F): 1.72 / Phase error: 28.16 / Stereochemistry target values: LS_WUNIT_K1 MOLECULAR REPLACEMENT / Resolution: 2.421→15.387 Å / SU ML: 0.33 / σ(F): 1.72 / Phase error: 28.16 / Stereochemistry target values: LS_WUNIT_K1Details: The sequence is d(CGGGCGCCCG). The molecule is placed in three-fold screw axis and form pseudo-continuous helix (Z-type) along c-axis. For this reason, the crystallographic asymmetric unit ...Details: The sequence is d(CGGGCGCCCG). The molecule is placed in three-fold screw axis and form pseudo-continuous helix (Z-type) along c-axis. For this reason, the crystallographic asymmetric unit consist of two tetramers such as d(CGCG) in 80% and d(GCGC) in 20%.
| |||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 172.268 Å2 / ksol: 0.6 e/Å3 | |||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.421→15.387 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| LS refinement shell | Highest resolution: 2.4213 Å
|
 Movie
Movie Controller
Controller








 PDBj
PDBj


