[English] 日本語
 Yorodumi
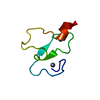
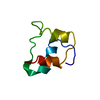
Yorodumi- PDB-3ci2: REFINEMENT OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF BARLEY ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3ci2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | REFINEMENT OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF BARLEY SERINE PROTEINASE INHIBITOR 2 AND COMPARISON WITH THE STRUCTURES IN CRYSTALS | ||||||
 Components Components | CHYMOTRYPSIN INHIBITOR 2 | ||||||
 Keywords Keywords | SERINE PROTEASE INHIBITOR | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Poulsen, F.M. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Refinement of the three-dimensional solution structure of barley serine proteinase inhibitor 2 and comparison with the structures in crystals. Authors: Ludvigsen, S. / Shen, H.Y. / Kjaer, M. / Madsen, J.C. / Poulsen, F.M. #1:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Accurate Measurements of Coupling Constants from Two-Dimensional Nuclear Magnetic Resonance Spectra of Proteins and Determination of Phi-Angles Authors: Ludvigsen, S. / Andersen, K.V. / Poulsen, F.M. #2:  Journal: J.Magn.Reson. / Year: 1990 Journal: J.Magn.Reson. / Year: 1990Title: Application of Symmetry Projection Operators to Measurements of Coupling Constants in 2D NMR Spectra of Proteins Authors: Shen, H. / Ludvigsen, S. / Poulsen, F.M. #3:  Journal: Alfred Benzon Symp. / Year: 1988 Journal: Alfred Benzon Symp. / Year: 1988Title: The Three-Dimensional Structure of Barley Serine Proteinase Inhibitor-2 in Solution as Determined by Proton Nuclear Magnetic Resonance Spectroscopy Authors: Poulsen, F.M. / Kjaer, M. / Clore, G.M. / Gronenborn, A.M. #4:  Journal: Protein Eng. / Year: 1987 Journal: Protein Eng. / Year: 1987Title: The Determination of the Three-Dimensional Structure of Barley Serine Proteinase Inhibitor 2 by Nuclear Magnetic Resonance, Distance Geometry and Restrained Molecular Dynamics Authors: Clore, G.M. / Gronenborn, A.M. / Kjaer, M. / Poulsen, F.M. #5:  Journal: Protein Eng. / Year: 1987 Journal: Protein Eng. / Year: 1987Title: Comparison of the Solution and X-Ray Structures of Barley Serine Proteinase Inhibitor 2 Authors: Clore, G.M. / Gronenborn, A.M. / James, M.N.G. / Kjaer, M. / Mcphalen, C.A. / Poulsen, F.M. #6:  Journal: Carlsberg Res.Commun. / Year: 1987 Journal: Carlsberg Res.Commun. / Year: 1987Title: Secondary Structure of Barley Serine Proteinase Inhibitor 2 Determined by Proton Nuclear Magnetic Resonance Spectroscopy Authors: Kjaer, M. / Poulsen, F.M. #7:  Journal: Carlsberg Res.Commun. / Year: 1987 Journal: Carlsberg Res.Commun. / Year: 1987Title: Sequence Specific Assignment of the Proton Nuclear Magnetic Resonance Spectrum of Barley Serine Proteinase Inhibitor 2 Authors: Kjaer, M. / Ludvigsen, S. / Sorensen, O.W. / Denys, L.A. / Kindtler, J. / Poulsen, F.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3ci2.cif.gz 3ci2.cif.gz | 424.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3ci2.ent.gz pdb3ci2.ent.gz | 352.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3ci2.json.gz 3ci2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ci/3ci2 https://data.pdbj.org/pub/pdb/validation_reports/ci/3ci2 ftp://data.pdbj.org/pub/pdb/validation_reports/ci/3ci2 ftp://data.pdbj.org/pub/pdb/validation_reports/ci/3ci2 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 7547.834 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| NMR ensemble | Conformers submitted total number: 20 |
|---|
 Movie
Movie Controller
Controller







 PDBj
PDBj