+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2ycm | ||||||
|---|---|---|---|---|---|---|---|
| Title | Inhibitors of herbicidal target IspD | ||||||
 Components Components | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC | ||||||
 Keywords Keywords | TRANSFERASE / HERBICIDE / ALLOSTERIC BINDING POCKET / NON-MEVALONATE-PATHWAY | ||||||
| Function / homology |  Function and homology information Function and homology information2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase / 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase activity / isopentenyl diphosphate biosynthetic process, methylerythritol 4-phosphate pathway / chloroplast stroma / chloroplast Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Hoeffken, H.W. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011Title: Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding. Authors: Witschel, M.C. / Hoeffken, H.W. / Seet, M. / Parra, L. / Mietzner, T. / Thater, F. / Niggeweg, R. / Rohl, F. / Illarionov, B. / Rohdich, F. / Kaiser, J. / Fischer, M. / Bacher, A. / Diederich, F. #1:  Journal: FEBS J. / Year: 2006 Journal: FEBS J. / Year: 2006Title: The Crystal Structure of a Plant 2C-Methyl-D- Erythritol 4-Phosphate Cytidylyltransferase Exhibits a Distinct Quaternary Structure Compared to Bacterial Homologues and a Possible Role in ...Title: The Crystal Structure of a Plant 2C-Methyl-D- Erythritol 4-Phosphate Cytidylyltransferase Exhibits a Distinct Quaternary Structure Compared to Bacterial Homologues and a Possible Role in Feedback Regulation for Cytidine Monophosphate. Authors: Gabrielsen, M. / Kaiser, J. / Rohdich, F. / Eisenreich, W. / Laupitz, R. / Bacher, A. / Bond, C.S. / Hunter, W.N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2ycm.cif.gz 2ycm.cif.gz | 104.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2ycm.ent.gz pdb2ycm.ent.gz | 79.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2ycm.json.gz 2ycm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2ycm_validation.pdf.gz 2ycm_validation.pdf.gz | 772.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2ycm_full_validation.pdf.gz 2ycm_full_validation.pdf.gz | 776.3 KB | Display | |
| Data in XML |  2ycm_validation.xml.gz 2ycm_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  2ycm_validation.cif.gz 2ycm_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yc/2ycm https://data.pdbj.org/pub/pdb/validation_reports/yc/2ycm ftp://data.pdbj.org/pub/pdb/validation_reports/yc/2ycm ftp://data.pdbj.org/pub/pdb/validation_reports/yc/2ycm | HTTPS FTP |
-Related structure data
| Related structure data |  2yc3SC  2yc5C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 25299.000 Da / Num. of mol.: 1 / Fragment: CYTIDYLTRANSFERASE DOMAIN, RESIDUES 76-302 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: P69834, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
|---|
-Non-polymers , 5 types, 99 molecules 


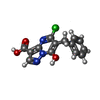





| #2: Chemical | | #3: Chemical | ChemComp-NA / | #4: Chemical | ChemComp-CU / | #5: Chemical | ChemComp-30A / | #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 48.48 % / Description: NONE |
|---|---|
| Crystal grow | Details: NATIVE CRYSTALS (SEE REMARK 1, PDB 1W77) WERE SOAKED WITH A SATURATED SOLUTION OF LIGAND |
-Data collection
| Diffraction | Mean temperature: 120 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06SA / Wavelength: 0.939272 / Beamline: X06SA / Wavelength: 0.939272 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Mar 16, 2006 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.939272 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→36.5 Å / Num. obs: 22629 / % possible obs: 99.8 % / Observed criterion σ(I): -3 / Redundancy: 6.9 % / Biso Wilson estimate: 38.6 Å2 / Rmerge(I) obs: 0.13 / Net I/σ(I): 10.84 |
| Reflection shell | Resolution: 1.8→1.9 Å / Redundancy: 6.7 % / Rmerge(I) obs: 1.36 / Mean I/σ(I) obs: 2.43 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2YC3 Resolution: 1.8→73.61 Å / Cor.coef. Fo:Fc: 0.951 / Cor.coef. Fo:Fc free: 0.944 / SU B: 6.984 / SU ML: 0.095 / Cross valid method: THROUGHOUT / ESU R: 0.205 / ESU R Free: 0.123 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES REFINED INDIVIDUALLY. THE ELECTRON DENSITY MAP SUGGESTS THAT IN THE CRYSTAL THE ALLOSTERIC POCKET IS ONLY PARTIALLY OCCUPIED AND ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. U VALUES REFINED INDIVIDUALLY. THE ELECTRON DENSITY MAP SUGGESTS THAT IN THE CRYSTAL THE ALLOSTERIC POCKET IS ONLY PARTIALLY OCCUPIED AND THEREFOR THE ELECTRON DENSITY REFLECTS AN OVERLAY OF THE UNLIGANDED AND LIGANDED ENZYME. THIS IS IN ACCORDANCE WITH THE HIGH TEMPERATURE FACTORS OF PART THE LIGAND AND OF THE AMINO ACIDS 264 TO 268 WHICH HAVE TO MOVE TO OPEN THE ALLOSTERIC BINDING POCKET.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.227 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→73.61 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




