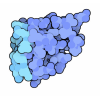
| Entry | Database: PDB / ID: 2wqp
|
|---|
| Title | Crystal structure of sialic acid synthase NeuB-inhibitor complex |
|---|
 Components Components | POLYSIALIC ACID CAPSULE BIOSYNTHESIS PROTEIN SIAC |
|---|
 Keywords Keywords | TRANSFERASE / NEUB / INHIBITOR / TIM BARREL / SIALIC ACID SYNTHASE |
|---|
| Function / homology |  Function and homology information Function and homology information
N-acetylneuraminic acid synthase, N-terminal / : / NeuB family / SAF domain / SAF / SAF domain / Type Iii Antifreeze Protein Isoform Hplc 12 / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal domain / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal / Antifreeze protein-like domain profile. ...N-acetylneuraminic acid synthase, N-terminal / : / NeuB family / SAF domain / SAF / SAF domain / Type Iii Antifreeze Protein Isoform Hplc 12 / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal domain / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal / Antifreeze protein-like domain profile. / Antifreeze-like/N-acetylneuraminic acid synthase C-terminal domain superfamily / Aldolase class I / Aldolase-type TIM barrel / TIM Barrel / Alpha-Beta Barrel / Alpha-Beta Complex / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |  NEISSERIA MENINGITIDIS (bacteria) NEISSERIA MENINGITIDIS (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / OTHER / Resolution: 1.75 Å SYNCHROTRON / OTHER / Resolution: 1.75 Å |
|---|
 Authors Authors | Liu, F. / Lee, H.J. / Strynadka, N.C.J. / Tanner, M.E. |
|---|
 Citation Citation |  Journal: Biochemistry / Year: 2009 Journal: Biochemistry / Year: 2009
Title: The Inhibition of Neisseria Meningitidis Sialic Acid Synthase by a Tetrahedral Intermediate Analog.
Authors: Liu, F. / Lee, H.J. / Strynadka, N.C.J. / Tanner, M.E. |
|---|
| History | | Deposition | Aug 25, 2009 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Sep 15, 2009 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jul 13, 2011 | Group: Advisory / Version format compliance |
|---|
| Revision 1.2 | Oct 12, 2011 | Group: Database references |
|---|
| Revision 1.3 | Nov 13, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Other / Structure summary
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_entry_details / pdbx_modification_feature / pdbx_struct_conn_angle / struct_conn / struct_site
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _pdbx_entry_details.has_protein_modification / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information NEISSERIA MENINGITIDIS (bacteria)
NEISSERIA MENINGITIDIS (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / OTHER / Resolution: 1.75 Å
SYNCHROTRON / OTHER / Resolution: 1.75 Å  Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2009
Journal: Biochemistry / Year: 2009 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2wqp.cif.gz
2wqp.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2wqp.ent.gz
pdb2wqp.ent.gz PDB format
PDB format 2wqp.json.gz
2wqp.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 2wqp_validation.pdf.gz
2wqp_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 2wqp_full_validation.pdf.gz
2wqp_full_validation.pdf.gz 2wqp_validation.xml.gz
2wqp_validation.xml.gz 2wqp_validation.cif.gz
2wqp_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/wq/2wqp
https://data.pdbj.org/pub/pdb/validation_reports/wq/2wqp ftp://data.pdbj.org/pub/pdb/validation_reports/wq/2wqp
ftp://data.pdbj.org/pub/pdb/validation_reports/wq/2wqp Links
Links Assembly
Assembly

 Components
Components NEISSERIA MENINGITIDIS (bacteria) / Strain: SEROGROUP B / Plasmid: PCWORI / Production host:
NEISSERIA MENINGITIDIS (bacteria) / Strain: SEROGROUP B / Plasmid: PCWORI / Production host: 










 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ALS
ALS  / Beamline: 4.2.2 / Wavelength: 0.979
/ Beamline: 4.2.2 / Wavelength: 0.979  Processing
Processing Movie
Movie Controller
Controller













 PDBj
PDBj