[English] 日本語
 Yorodumi
Yorodumi- PDB-2o8k: NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promote... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2o8k | ||||||
|---|---|---|---|---|---|---|---|
| Title | NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION/DNA / PROTEIN-DNA COMPLEX / HELIX-TURN-HELIX / TRANSCRIPTION FACTOR / SIGMA-54 / RNA POLYMERASE / TRANSCRIPTION-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA-binding transcription activator activity / sigma factor activity / nucleotidyltransferase activity / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / protein-DNA complex / transcription cis-regulatory region binding / regulation of DNA-templated transcription Similarity search - Function | ||||||
| Biological species |   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | ||||||
| Method | SOLUTION NMR / docking, rigid-body minimization, simulated annealing | ||||||
 Authors Authors | Doucleff, M. / Pelton, J.G. / Lee, P.S. / Wemmer, D.E. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2007 Journal: J.Mol.Biol. / Year: 2007Title: Structural basis of DNA recognition by the alternative sigma-factor, sigma54. Authors: Doucleff, M. / Pelton, J.G. / Lee, P.S. / Nixon, B.T. / Wemmer, D.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2o8k.cif.gz 2o8k.cif.gz | 766.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2o8k.ent.gz pdb2o8k.ent.gz | 639.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2o8k.json.gz 2o8k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/o8/2o8k https://data.pdbj.org/pub/pdb/validation_reports/o8/2o8k ftp://data.pdbj.org/pub/pdb/validation_reports/o8/2o8k ftp://data.pdbj.org/pub/pdb/validation_reports/o8/2o8k | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 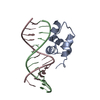
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 4252.761 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 4306.846 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: Protein | Mass: 7488.800 Da / Num. of mol.: 1 / Fragment: C-TERMINAL RPON DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Aquifex aeolicus (bacteria) / Gene: rpoN / Plasmid: pSKB3 / Production host: Aquifex aeolicus (bacteria) / Gene: rpoN / Plasmid: pSKB3 / Production host:  Strain (production host): BL21(DE3) with Rosetta.pLysS plasmid References: UniProt: O66858 |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: docking, rigid-body minimization, simulated annealing Software ordinal: 1 Details: protein and DNA structures determined individually via simulated annealing; the two structures were docked using rigid body minimization; the whole structure was refined by simulated annealing | ||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: 20 structures with lowest energy and no restraint violations greater than 0.5 A and 5 degrees for distance and dihedral restraints, respectively Conformers calculated total number: 50 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller













 PDBj
PDBj






































 NMRPipe
NMRPipe