[English] 日本語
 Yorodumi
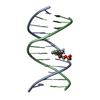
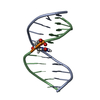
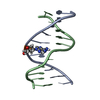
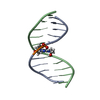
Yorodumi- PDB-2mnx: Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mnx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / B-form / 11-mer | ||||||
| Function / homology | DNA / DNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
| Model details | closest to the average, model1 | ||||||
 Authors Authors | Kowal, E.A. / Kotapati, S. / Turo, M. / Tretyakova, N. / Stone, M.P. | ||||||
 Citation Citation |  Journal: Chem.Res.Toxicol. / Year: 2014 Journal: Chem.Res.Toxicol. / Year: 2014Title: Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene. Authors: Kowal, E.A. / Wickramaratne, S. / Kotapati, S. / Turo, M. / Tretyakova, N. / Stone, M.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mnx.cif.gz 2mnx.cif.gz | 133.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mnx.ent.gz pdb2mnx.ent.gz | 106.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mnx.json.gz 2mnx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mn/2mnx https://data.pdbj.org/pub/pdb/validation_reports/mn/2mnx ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mnx ftp://data.pdbj.org/pub/pdb/validation_reports/mn/2mnx | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3486.353 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 3291.145 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||||
| NMR constraints | NA alpha-angle constraints total count: 18 / NA beta-angle constraints total count: 18 / NA delta-angle constraints total count: 18 / NA epsilon-angle constraints total count: 18 / NA gamma-angle constraints total count: 18 / NOE constraints total: 283 / NOE intraresidue total count: 125 / NOE sequential total count: 158 | ||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: back calculated data agree with experimental NOESY spectrum Conformers calculated total number: 9 / Conformers submitted total number: 9 |
 Movie
Movie Controller
Controller





 PDBj
PDBj






































 Amber
Amber