+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mer | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of helix 69 from escherichia coli 23s ribosomal rna | ||||||||||||||||||||
 Components Components | RNA_(5'-R(P* Keywords KeywordsRNA / Pseudouridine / Helix 69 / Ribosomal | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing, molecular dynamics, torsion angle dynamics | Model details | lowest energy, model1 |  Authors AuthorsJiang, J. / Aduri, R. / Chow, C.S. / Santalucia, J. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2014 Journal: Nucleic Acids Res. / Year: 2014Title: Structure modulation of helix 69 from Escherichia coli 23S ribosomal RNA by pseudouridylations. Authors: Jiang, J. / Aduri, R. / Chow, C.S. / Santalucia, J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mer.cif.gz 2mer.cif.gz | 122.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mer.ent.gz pdb2mer.ent.gz | 100.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mer.json.gz 2mer.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2mer_validation.pdf.gz 2mer_validation.pdf.gz | 343.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2mer_full_validation.pdf.gz 2mer_full_validation.pdf.gz | 399.3 KB | Display | |
| Data in XML |  2mer_validation.xml.gz 2mer_validation.xml.gz | 9.3 KB | Display | |
| Data in CIF |  2mer_validation.cif.gz 2mer_validation.cif.gz | 13.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/me/2mer https://data.pdbj.org/pub/pdb/validation_reports/me/2mer ftp://data.pdbj.org/pub/pdb/validation_reports/me/2mer ftp://data.pdbj.org/pub/pdb/validation_reports/me/2mer | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 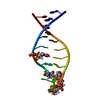
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 6077.673 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, molecular dynamics, torsion angle dynamics Software ordinal: 1 | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: CONVERGED LOWEST ENERGY STRUCTURES Conformers calculated total number: 100 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller








 PDBj
PDBj





























