[English] 日本語
 Yorodumi
Yorodumi- PDB-2m21: Solution structure of the Tetrahymena telomerase RNA stem IV term... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2m21 | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | |||||||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / holoenzyme / protozoan proteins / ciliate / template / genetic / reverse transcriptase | Function / homology | RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing | Model details | lowest energy, model1 |  Authors AuthorsCash, D.D. / Richards, R.J. / Wu, H. / Trantirek, L. / O'Connor, C.M. / Feigon, J. / Collins, K. |  Citation Citation Journal: Rna / Year: 2006 Journal: Rna / Year: 2006Title: Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. Authors: Richards, R.J. / Wu, H. / Trantirek, L. / O'Connor, C.M. / Collins, K. / Feigon, J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2m21.cif.gz 2m21.cif.gz | 184.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2m21.ent.gz pdb2m21.ent.gz | 152.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2m21.json.gz 2m21.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/m2/2m21 https://data.pdbj.org/pub/pdb/validation_reports/m2/2m21 ftp://data.pdbj.org/pub/pdb/validation_reports/m2/2m21 ftp://data.pdbj.org/pub/pdb/validation_reports/m2/2m21 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 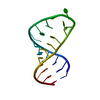
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 6649.980 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 / Details: residual polar couplings | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 14 |
 Movie
Movie Controller
Controller







 PDBj
PDBj





























 X-PLOR NIH
X-PLOR NIH