[English] 日本語
 Yorodumi
Yorodumi- PDB-2k71: Structure and dynamics of a DNA GNRA hairpin solved vy high-sensi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2k71 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and dynamics of a DNA GNRA hairpin solved vy high-sensitivity NMR with two independent converging methods, simulated annealing (DYANA) and mesoscopic molecular modelling (BCE/AMBER) | ||||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / tetraloop hairpin / B-DNA-like / 2 non canonical torsions / unusual BIIz+ torsions | Function / homology | DNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing, Biopolymer Chain Elasticity approach | Model details | DNA GNRA tetraloop structure |  Authors AuthorsSantini, G.P.H. / Cognet, J.A.H. / Xu, D. / Singarapu, K.K. / Herve du Penhoat, C.L.M. |  Citation Citation Journal: J.Phys.Chem.B / Year: 2009 Journal: J.Phys.Chem.B / Year: 2009Title: Nucleic acid folding determined by mesoscale modeling and NMR spectroscopy: solution structure of d(GCGAAAGC). Authors: Santini, G.P. / Cognet, J.A. / Xu, D. / Singarapu, K.K. / Herve du Penhoat, C. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2k71.cif.gz 2k71.cif.gz | 89.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2k71.ent.gz pdb2k71.ent.gz | 60.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2k71.json.gz 2k71.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k7/2k71 https://data.pdbj.org/pub/pdb/validation_reports/k7/2k71 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/2k71 ftp://data.pdbj.org/pub/pdb/validation_reports/k7/2k71 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 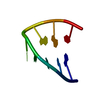
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2460.644 Da / Num. of mol.: 1 / Fragment: DNA GNRA hairpin / Source method: obtained synthetically |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR / Details: DNA GNRA tetraloop structure | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, Biopolymer Chain Elasticity approach Software ordinal: 1 Details: 100 structures were used in DYANA simulated annealing approach to give models 1 to 20 that were energy-refined with AMBER, Model number 21 was constructed with the BCE approach in agreement ...Details: 100 structures were used in DYANA simulated annealing approach to give models 1 to 20 that were energy-refined with AMBER, Model number 21 was constructed with the BCE approach in agreement with the NMR data and then energy-refined with AMBER | ||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 101 / Conformers submitted total number: 21 |
 Movie
Movie Controller
Controller








 PDBj
PDBj

 HSQC
HSQC