[English] 日本語
 Yorodumi
Yorodumi- PDB-2hpq: Structures of the noncovalent complexes of human and bovine proth... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2hpq | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human ppack-thrombin | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / SERINE PROTEINASE / HYDROLASE-HYDROLASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information: / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium / positive regulation of phospholipase C-activating G protein-coupled receptor signaling pathway / Defective F8 cleavage by thrombin ...: / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium / positive regulation of phospholipase C-activating G protein-coupled receptor signaling pathway / Defective F8 cleavage by thrombin / Platelet Aggregation (Plug Formation) / ligand-gated ion channel signaling pathway / positive regulation of collagen biosynthetic process / negative regulation of platelet activation / negative regulation of blood coagulation / positive regulation of blood coagulation / negative regulation of fibrinolysis / regulation of cytosolic calcium ion concentration / Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus / Gamma-carboxylation of protein precursors / Common Pathway of Fibrin Clot Formation / Removal of aminoterminal propeptides from gamma-carboxylated proteins / fibrinolysis / Intrinsic Pathway of Fibrin Clot Formation / negative regulation of proteolysis / negative regulation of cytokine production involved in inflammatory response / Peptide ligand-binding receptors / Regulation of Complement cascade / positive regulation of release of sequestered calcium ion into cytosol / acute-phase response / Cell surface interactions at the vascular wall / positive regulation of receptor signaling pathway via JAK-STAT / growth factor activity / lipopolysaccharide binding / positive regulation of insulin secretion / platelet activation / response to wounding / positive regulation of protein localization to nucleus / Golgi lumen / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / positive regulation of reactive oxygen species metabolic process / blood coagulation / antimicrobial humoral immune response mediated by antimicrobial peptide / regulation of cell shape / heparin binding / : / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cell growth / blood microparticle / G alpha (q) signalling events / cell surface receptor signaling pathway / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / receptor ligand activity / endoplasmic reticulum lumen / signaling receptor binding / serine-type endopeptidase activity / positive regulation of cell population proliferation / calcium ion binding / proteolysis / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Homo Sapiens (human) Homo Sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 3.3 Å X-RAY DIFFRACTION / Resolution: 3.3 Å | ||||||
 Authors Authors | Tulinsky, A. / Padmanabhan, K. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1993 Journal: Biochemistry / Year: 1993Title: Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin. Authors: Arni, R.K. / Padmanabhan, K. / Padmanabhan, K.P. / Wu, T.P. / Tulinsky, A. #1:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Structure of the Hirugen and Hirulog 1 Complexes of Alpha-Thrombin Authors: Skrzypczak-Jankun, E. / Carperos, V.E. / Ravichandran, K.G. / Tulinsky, A. / Westbrook, M. / Maraganore, J.M. #2:  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: The Refined Structure of the Epsilon-Aminocaproic Acid Complex of Human Plasminogen Kringle 4 Authors: Wu, T.-P. / Padmanabhan, K. / Tulinsky, A. / Mulichak, A.M. #3:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Structure of Bovine Prothrombin Fragment 1 Refined at 2.25 Angstroms Resolution Authors: Seshadri, T.P. / Tulinsky, A. / Skrzypczak-Jankun, E. / Park, C.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2hpq.cif.gz 2hpq.cif.gz | 91.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2hpq.ent.gz pdb2hpq.ent.gz | 67.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2hpq.json.gz 2hpq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hp/2hpq https://data.pdbj.org/pub/pdb/validation_reports/hp/2hpq ftp://data.pdbj.org/pub/pdb/validation_reports/hp/2hpq ftp://data.pdbj.org/pub/pdb/validation_reports/hp/2hpq | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 37 / 2: RESIDUE PHE I 1 IS A D-AMINO ACID. |
- Components
Components
| #1: Protein/peptide | Mass: 4096.534 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P00734, thrombin Homo sapiens (human) / References: UniProt: P00734, thrombin | ||
|---|---|---|---|
| #2: Protein | Mass: 29780.219 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P00734, thrombin Homo sapiens (human) / References: UniProt: P00734, thrombin | ||
| #3: Protein | Mass: 8692.646 Da / Num. of mol.: 1 / Fragment: Activation peptide fragment 2 / Source method: isolated from a natural source / Source: (natural)  Homo Sapiens (human) / References: UniProt: P00734, thrombin Homo Sapiens (human) / References: UniProt: P00734, thrombin | ||
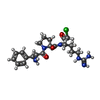
| #4: Chemical | ChemComp-0G7 / | ||
| #5: Water | ChemComp-HOH / | ||
| Compound details | THROMBIN IS CLEAVED BETWEEN RESIDUES 15 AND 16. CHAIN INDICATOR *L* IS USED FOR RESIDUES 1H - 15 ...THROMBIN IS CLEAVED BETWEEN RESIDUES 15 AND 16. CHAIN INDICATOR *L* IS USED FOR RESIDUES 1H - 15 AND CHAIN INDICATOR *H* IS USED FOR RESIDUES 16 - 247. CHAIN INDICATOR *P* IS USED FOR PROTHROMBI | ||
| Has protein modification | Y | ||
| Nonpolymer details | THE INHIBITOR IS COVALENTLY| Sequence details | CHYMOTRYPSIN NUMBERING (RATHER THAN SEQUENTIAL) SYSTEM IS USED, BASED ON THE TOPOLOGICAL ALIGNMENT ...CHYMOTRYPS | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.49 Å3/Da / Density % sol: 72.6 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 60 % | ||||||||||||||||||||
| Crystal grow | *PLUS pH: 5.5 / Method: vapor diffusion, hanging drop / Details: using macroseeding | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 3.2 Å / Num. obs: 9195 / Observed criterion σ(I): 2 / Num. measured all: 51004 / Rmerge(I) obs: 0.065 |
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor obs: 0.155 / Highest resolution: 3.3 Å Details: A FEW SIDE CHAINS IN BOTH THROMBIN AND FRAGMENT 2 DO NOT HAVE WELL DEFINED ELECTRON DENSITY. THESE ATOMS HAVE BEEN GIVEN OCCUPANCIES OF 0.0 IN THE FILE. THE FOLLOWING RESIDUES IN FRAGMENT 2 ...Details: A FEW SIDE CHAINS IN BOTH THROMBIN AND FRAGMENT 2 DO NOT HAVE WELL DEFINED ELECTRON DENSITY. THESE ATOMS HAVE BEEN GIVEN OCCUPANCIES OF 0.0 IN THE FILE. THE FOLLOWING RESIDUES IN FRAGMENT 2 DO NOT HAVE ELECTRON DENSITY FOR THE SIDE CHAIN BEYOND CB: VAL 302, ARG 305, GLN 307, GLN 310, ARG 312, THR 316, SER 327, LYS 331, SER 341, VAL 343, GLN 344, VAL 346, LYS 367 AND ASN 377. IN ADDITION, THERE WAS NO ELECTRON DENSITY FOR THE 14 N-TERMINAL AND 23 C-TERMINAL INTERKRINGLE PEPTIDES. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 3.3 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 3.3 Å / Lowest resolution: 10 Å / Rfactor obs: 0.155 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 19 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 3.3 Å / Lowest resolution: 3.6 Å / Num. reflection Rfree: 1542 / Rfactor obs: 0.157 |
 Movie
Movie Controller
Controller













 PDBj
PDBj