[English] 日本語
 Yorodumi
Yorodumi- PDB-2h3o: Structure of MERFT, a membrane protein with two trans-membrane helices -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2h3o | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of MERFT, a membrane protein with two trans-membrane helices | ||||||
 Components Components | MerF | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / ALPHA-HELIX / BICELLE | ||||||
| Function / homology | bacterial mercury transporter, merf / Mercury ion transport, MerF / Membrane transport protein MerF / Helix Hairpins / Orthogonal Bundle / Mainly Alpha / membrane / MerF Function and homology information Function and homology information | ||||||
| Biological species |  Morganella morganii (bacteria) Morganella morganii (bacteria) | ||||||
| Method | SOLID-STATE NMR / DIRECT STRUCTURAL FITTING OF 2D SOLID-STATE NMR DATA | ||||||
 Authors Authors | Opella, S.J. / De Angelis, A.A. / Howell, S.C. / Nevzorov, A.A. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2006 Journal: J.Am.Chem.Soc. / Year: 2006Title: Structure Determination of a Membrane Protein with Two Trans-membrane Helices in Aligned Phospholipid Bicelles by Solid-State NMR Spectroscopy. Authors: De Angelis, A.A. / Howell, S.C. / Nevzorov, A.A. / Opella, S.J. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE Residue at position 72 is a C72S mutation. Residue Ser is further modified to Homoserine |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2h3o.cif.gz 2h3o.cif.gz | 15.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2h3o.ent.gz pdb2h3o.ent.gz | 6.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2h3o.json.gz 2h3o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2h3o_validation.pdf.gz 2h3o_validation.pdf.gz | 246.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2h3o_full_validation.pdf.gz 2h3o_full_validation.pdf.gz | 246.6 KB | Display | |
| Data in XML |  2h3o_validation.xml.gz 2h3o_validation.xml.gz | 2 KB | Display | |
| Data in CIF |  2h3o_validation.cif.gz 2h3o_validation.cif.gz | 2.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/h3/2h3o https://data.pdbj.org/pub/pdb/validation_reports/h3/2h3o ftp://data.pdbj.org/pub/pdb/validation_reports/h3/2h3o ftp://data.pdbj.org/pub/pdb/validation_reports/h3/2h3o | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 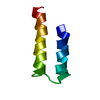
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6408.632 Da / Num. of mol.: 1 / Fragment: Helix-loop-helix, residues 12-72 / Mutation: C21S, C22S, C71S, C72(GLX) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Morganella morganii (bacteria) / Gene: merF / Plasmid: PET31B-MERFT / Production host: Morganella morganii (bacteria) / Gene: merF / Plasmid: PET31B-MERFT / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLID-STATE NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||
| NMR details | Text: 15N chemical shifts and 1H-15N dipolar couplings were measured by solid-state NMR |
- Sample preparation
Sample preparation
| Details | Contents: 6 mM MerFt aligned in large 14-O-PC/6-O-PC phospholipid bicelles, 28% (w/v) in H2O Solvent system: H2O |
|---|---|
| Sample conditions | pH: 5 / Pressure: ambient / Temperature: 313 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker AVANCE / Manufacturer: Bruker / Model: AVANCE / Field strength: 700 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DIRECT STRUCTURAL FITTING OF 2D SOLID-STATE NMR DATA Software ordinal: 1 Details: This structure was calculated by using a structural fitting algorithm that finds torsion angles between consecutive residues based on their 15N chemical shift, 1H-15N dipolar coupling ...Details: This structure was calculated by using a structural fitting algorithm that finds torsion angles between consecutive residues based on their 15N chemical shift, 1H-15N dipolar coupling frequencies and Ramachandran maps. One hydrophobic matching and one loose helix-helix distance restraints were used | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 20 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller






 PDBj
PDBj NMRPipe
NMRPipe