[English] 日本語
 Yorodumi
Yorodumi- PDB-2gun: RNA-Binding Affinities and Crystal Structure of Oligonucleotides ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2gun | ||||||
|---|---|---|---|---|---|---|---|
| Title | RNA-Binding Affinities and Crystal Structure of Oligonucleotides containing Five-Atom Amide-Based Backbone Structures | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA/RNA / DNA:RNA hybrid / amide bearing DNA / thymidine dimers / internucleosidic amide linkage / DNA / RNA / DNA-RNA COMPLEX | ||||||
| Function / homology | COBALT (III) ION / SPERMINE / DNA / RNA Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Pallan, P.S. / Egli, M. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2006 Journal: Biochemistry / Year: 2006Title: RNA-Binding Affinities and Crystal Structure of Oligonucleotides Containing Five-Atom Amide-Based Backbone Structures. Authors: Pallan, P.S. / von Matt, P. / Wilds, C.J. / Altmann, K.-H. / Egli, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2gun.cif.gz 2gun.cif.gz | 21.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2gun.ent.gz pdb2gun.ent.gz | 13.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2gun.json.gz 2gun.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gu/2gun https://data.pdbj.org/pub/pdb/validation_reports/gu/2gun ftp://data.pdbj.org/pub/pdb/validation_reports/gu/2gun ftp://data.pdbj.org/pub/pdb/validation_reports/gu/2gun | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 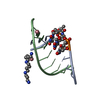
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 2745.793 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||||
|---|---|---|---|---|---|
| #2: RNA chain | Mass: 2965.895 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||||
| #3: Chemical | | #4: Chemical | ChemComp-SPM / | #5: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.22 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 0.5 mM cobalt hexamine, 5 mM spermine tetrachloride, 80 mM sodium cacodylate buffer (pH = 7.0) and 10% (v/v) 2-methyl-2,4-pentanediol (MPD)., VAPOR DIFFUSION, HANGING DROP, temperature 291K | ||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 110 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 5ID-B / Wavelength: 0.9203 Å / Beamline: 5ID-B / Wavelength: 0.9203 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Nov 4, 2001 |
| Radiation | Monochromator: Si Monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9203 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→20 Å / Num. all: 2602 / Num. obs: 2391 / % possible obs: 91.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Rsym value: 0.057 |
| Reflection shell | Highest resolution: 2.8 Å / % possible all: 92.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 2.8→20 Å / Cross valid method: THROUGHOUT / σ(F): 4 MOLECULAR REPLACEMENT / Resolution: 2.8→20 Å / Cross valid method: THROUGHOUT / σ(F): 4 Stereochemistry target values: L. Clowney et.al., Geometric Parameters in Nucleic Acids: Nitrogenous Bases J. Am. Chem. Soc., (1996) 118, 509-518; A. Gelbin et.al., Geometric Parameters in Nucleic ...Stereochemistry target values: L. Clowney et.al., Geometric Parameters in Nucleic Acids: Nitrogenous Bases J. Am. Chem. Soc., (1996) 118, 509-518; A. Gelbin et.al., Geometric Parameters in Nucleic Acids: Sugar and Phosphate Constituents J. Am. Chem. Soc., (1996) 118, 519-529. G. Parkinson et. al., New parameters for the refinement of nucleic acid-containing structures Acta Cryst. (1996). D52, 57-64.
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→20 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller








 PDBj
PDBj







