+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2gk2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the N terminal domain of human CEACAM1 | ||||||
 Components Components | Carcinoembryonic antigen-related cell adhesion molecule 1 | ||||||
 Keywords Keywords | CELL ADHESION / immunoglobulin domain / adhesion protein | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of endothelial cell differentiation / insulin receptor internalization / negative regulation of cytotoxic T cell degranulation / granulocyte colony-stimulating factor signaling pathway / regulation of homophilic cell adhesion / Regulation of MITF-M dependent genes involved in invasion / negative regulation of hepatocyte proliferation / regulation of epidermal growth factor receptor signaling pathway / regulation of sprouting angiogenesis / filamin binding ...regulation of endothelial cell differentiation / insulin receptor internalization / negative regulation of cytotoxic T cell degranulation / granulocyte colony-stimulating factor signaling pathway / regulation of homophilic cell adhesion / Regulation of MITF-M dependent genes involved in invasion / negative regulation of hepatocyte proliferation / regulation of epidermal growth factor receptor signaling pathway / regulation of sprouting angiogenesis / filamin binding / regulation of blood vessel remodeling / negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of lipid biosynthetic process / negative regulation of T cell mediated cytotoxicity / regulation of endothelial cell migration / negative regulation of granulocyte differentiation / insulin catabolic process / regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / common myeloid progenitor cell proliferation / Fibronectin matrix formation / : / negative regulation of interleukin-1 production / positive regulation of vasculogenesis / negative regulation of fatty acid biosynthetic process / negative regulation of platelet aggregation / bile acid transmembrane transporter activity / negative regulation of vascular permeability / regulation of immune system process / wound healing, spreading of cells / negative regulation of T cell receptor signaling pathway / bile acid and bile salt transport / transport vesicle membrane / blood vessel development / microvillus membrane / homophilic cell-cell adhesion / tertiary granule membrane / lateral plasma membrane / regulation of ERK1 and ERK2 cascade / negative regulation of protein kinase activity / specific granule membrane / regulation of cell migration / protein tyrosine kinase binding / basal plasma membrane / Cell surface interactions at the vascular wall / integrin-mediated signaling pathway / adherens junction / regulation of cell growth / kinase binding / cellular response to insulin stimulus / cell-cell junction / cell junction / cell migration / actin binding / angiogenesis / protein phosphatase binding / calmodulin binding / cell adhesion / protein dimerization activity / apical plasma membrane / Neutrophil degranulation / cell surface / signal transduction / protein homodimerization activity / extracellular exosome / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Fedarovich, A. / Tomberg, J. / Nicholas, R.A. / Davies, C. | ||||||
 Citation Citation |  Journal: Acta Crystallogr.,Sect.D / Year: 2006 Journal: Acta Crystallogr.,Sect.D / Year: 2006Title: Structure of the N-terminal domain of human CEACAM1: binding target of the opacity proteins during invasion of Neisseria meningitidis and N. gonorrhoeae. Authors: Fedarovich, A. / Tomberg, J. / Nicholas, R.A. / Davies, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2gk2.cif.gz 2gk2.cif.gz | 58.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2gk2.ent.gz pdb2gk2.ent.gz | 42 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2gk2.json.gz 2gk2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gk/2gk2 https://data.pdbj.org/pub/pdb/validation_reports/gk/2gk2 ftp://data.pdbj.org/pub/pdb/validation_reports/gk/2gk2 ftp://data.pdbj.org/pub/pdb/validation_reports/gk/2gk2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1l6zS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 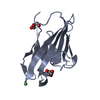
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 12171.431 Da / Num. of mol.: 2 / Fragment: N terminal domain (residues 63-170) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CEACAM1 / Plasmid: pGEX-2V / Production host: Homo sapiens (human) / Gene: CEACAM1 / Plasmid: pGEX-2V / Production host:  #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.73 Å3/Da / Density % sol: 54.95 % |
|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: 16% (w/v) polyethylene glycol mono methyl ether 2000, 100 mM Tris, and 5 mM nickel chloride, pH 8.5, VAPOR DIFFUSION, HANGING DROP, temperature 294K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-ID / Wavelength: 1 Å / Beamline: 22-ID / Wavelength: 1 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Jun 26, 2005 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→50 Å / Num. all: 13366 / Num. obs: 13366 / % possible obs: 99.6 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 10.5 % / Biso Wilson estimate: 50 Å2 / Rmerge(I) obs: 0.067 / Net I/σ(I): 38.8 |
| Reflection shell | Resolution: 2.2→2.28 Å / Redundancy: 6.7 % / Rmerge(I) obs: 0.736 / Mean I/σ(I) obs: 2 / Num. unique all: 1290 / % possible all: 96.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1L6Z Resolution: 2.2→50 Å / Cor.coef. Fo:Fc: 0.941 / Cor.coef. Fo:Fc free: 0.909 / SU B: 11.806 / SU ML: 0.152 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.271 / ESU R Free: 0.218 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 41.5 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→50 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.257 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj










