[English] 日本語
 Yorodumi
Yorodumi- PDB-2fr3: Crystal Structure of Cellular Retinoic Acid Binding Protein Type ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2fr3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Cellular Retinoic Acid Binding Protein Type II in Complex with All-Trans-Retinoic Acid at 1.48 Angstroms Resolution | ||||||
 Components Components | Cellular retinoic acid binding protein 2 | ||||||
 Keywords Keywords | TRANSPORT PROTEIN / CRABPII / Retinoic Acid / Retinoids / Beta Barrel / High Resolution | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of collateral sprouting / retinoid binding / retinoic acid binding / retinal binding / embryonic forelimb morphogenesis / retinoic acid metabolic process / retinol binding / Signaling by Retinoic Acid / epidermis development / fatty acid transport ...positive regulation of collateral sprouting / retinoid binding / retinoic acid binding / retinal binding / embryonic forelimb morphogenesis / retinoic acid metabolic process / retinol binding / Signaling by Retinoic Acid / epidermis development / fatty acid transport / cyclin binding / fatty acid binding / regulation of DNA-templated transcription / endoplasmic reticulum / signal transduction / extracellular exosome / nucleoplasm / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Rigid Body Refinement / Resolution: 1.48 Å SYNCHROTRON / Rigid Body Refinement / Resolution: 1.48 Å | ||||||
 Authors Authors | Vaezeslami, S. / Geiger, J.H. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2006 Journal: J.Mol.Biol. / Year: 2006Title: The structure of Apo-wild-type cellular retinoic acid binding protein II at 1.4 A and its relationship to ligand binding and nuclear translocation. Authors: Vaezeslami, S. / Mathes, E. / Vasileiou, C. / Borhan, B. / Geiger, J.H. #1:  Journal: Thesis Journal: ThesisTitle: Determining crystal structures of proteins and protein complexes by X-ray crystallography: X-ray crystallographic studies of the mutants of cellular retinoic acid binding protein type II ...Title: Determining crystal structures of proteins and protein complexes by X-ray crystallography: X-ray crystallographic studies of the mutants of cellular retinoic acid binding protein type II toward designing a mimic of rhodopsin Authors: Vaezeslami, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2fr3.cif.gz 2fr3.cif.gz | 78.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2fr3.ent.gz pdb2fr3.ent.gz | 59.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2fr3.json.gz 2fr3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fr/2fr3 https://data.pdbj.org/pub/pdb/validation_reports/fr/2fr3 ftp://data.pdbj.org/pub/pdb/validation_reports/fr/2fr3 ftp://data.pdbj.org/pub/pdb/validation_reports/fr/2fr3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2frsC 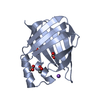 2fs6C  2fs7C  2fru C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15581.802 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CRABP2 / Plasmid: pET17-b / Production host: Homo sapiens (human) / Gene: CRABP2 / Plasmid: pET17-b / Production host:  |
|---|---|
| #2: Chemical | ChemComp-ACT / |
| #3: Chemical | ChemComp-REA / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.55 Å3/Da / Density % sol: 51.77 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5.4 Details: 30% (w/v) PEG 4000, 0.1 M sodium citrate/citric acid pH 5.4, and 0.2 ammonium acetate (crystallization performed in dark room and under red light), VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 32-ID / Wavelength: 1 Å / Beamline: 32-ID / Wavelength: 1 Å | |||||||||||||||
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Jul 23, 2005 | |||||||||||||||
| Radiation |
| |||||||||||||||
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 | |||||||||||||||
| Reflection | Resolution: 1.48→39.81 Å / Num. all: 27332 / Num. obs: 26090 / % possible obs: 95.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Biso Wilson estimate: 17.1 Å2 / Rmerge(I) obs: 0.044 / Χ2: 2.781 / Net I/σ(I): 28.19 | |||||||||||||||
| Reflection shell | Resolution: 1.48→1.53 Å / % possible obs: 96.4 % / Rmerge(I) obs: 0.139 / Mean I/σ(I) obs: 5.08 / Num. unique obs: 2590 / Χ2: 0.777 / % possible all: 96.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: Rigid Body Refinement / Resolution: 1.48→39.81 Å / Cor.coef. Fo:Fc: 0.977 / Cor.coef. Fo:Fc free: 0.96 / SU B: 1.952 / SU ML: 0.034 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.068 / ESU R Free: 0.065 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.735 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.48→39.81 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.48→1.515 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj