[English] 日本語
 Yorodumi
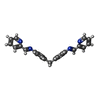
Yorodumi- PDB-2et0: The structure of a three-way DNA junction in complex with a metal... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2et0 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | ||||||||||||||||||||
 Components Components | 5'-D(* Keywords KeywordsDNA / Drug-DNA complex / 3-way junction / DNA structure recognition | Function / homology | : / Chem-NPM / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.7 Å MAD / Resolution: 1.7 Å  Authors AuthorsOleksi, A. / Blanco, A.G. / Boer, R. / Uson, I. / Aymami, J. / Coll, M. |  Citation Citation Journal: ANGEW.CHEM.INT.ED.ENGL. / Year: 2006 Journal: ANGEW.CHEM.INT.ED.ENGL. / Year: 2006Title: Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate Authors: Oleksi, A. / Blanco, A.G. / Boer, R. / Uson, I. / Aymami, J. / Rodger, A. / Hannon, M.J. / Coll, M. History |
Remark 600 | HETEROGEN The het group NPM 1, 2, 3 exists as a complex of three molecules of NPM with two FE(II) ...HETEROGEN The het group NPM 1, 2, 3 exists as a complex of three molecules of NPM with two FE(II) molecules in the structure. The IUPAC name for the complete ligand is 'tris (1,1-bis(N-(4-phenyl)-2-pyridylcaboxaldimine) methane) bis-iron(II) tetrachloride | |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2et0.cif.gz 2et0.cif.gz | 26.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2et0.ent.gz pdb2et0.ent.gz | 17.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2et0.json.gz 2et0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2et0_validation.pdf.gz 2et0_validation.pdf.gz | 1.4 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2et0_full_validation.pdf.gz 2et0_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  2et0_validation.xml.gz 2et0_validation.xml.gz | 4.6 KB | Display | |
| Data in CIF |  2et0_validation.cif.gz 2et0_validation.cif.gz | 5.6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/et/2et0 https://data.pdbj.org/pub/pdb/validation_reports/et/2et0 ftp://data.pdbj.org/pub/pdb/validation_reports/et/2et0 ftp://data.pdbj.org/pub/pdb/validation_reports/et/2et0 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 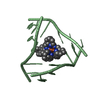
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
| ||||||||||||
| Details | The asymmetric unit contains one-third of the 3-way junction, the 2nd and 3rd part are generated by the cube body diagonal 3-fold axis. For residues 10 to 16, the biological assembly is constructed by applying symops z,x,y and y,z,x |
- Components
Components
| #1: DNA chain | Mass: 1809.217 Da / Num. of mol.: 2 / Source method: obtained synthetically #2: Chemical | ChemComp-FE2 / #3: Chemical | ChemComp-NPM / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.98 Å3/Da / Density % sol: 69.1 % | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 0.08M Mg acetate, 0.05M TrisCl, pH 8.5, 5% PEG400, VAPOR DIFFUSION, SITTING DROP, temperature 298.0K | ||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||
| Detector |
| ||||||||||||||||||
| Radiation |
| ||||||||||||||||||
| Radiation wavelength |
| ||||||||||||||||||
| Reflection | Resolution: 1.7→22.8 Å / Num. all: 7149 / Num. obs: 7149 / % possible obs: 99.2 % / Observed criterion σ(I): -3 / Redundancy: 50 % / Rmerge(I) obs: 0.0537 / Net I/σ(I): 29.1 | ||||||||||||||||||
| Reflection shell | Highest resolution: 1.7 Å / % possible all: 97.8 |
- Processing
Processing
| Software |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 1.7→22.5 Å / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 4 / σ(I): 4 / Stereochemistry target values: Engh & Huber MAD / Resolution: 1.7→22.5 Å / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 4 / σ(I): 4 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||
| Displacement parameters | Biso mean: 23.45 Å2 | |||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→22.5 Å
| |||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj