[English] 日本語
 Yorodumi
Yorodumi- PDB-2d09: A Role for Active Site Water Molecules and Hydroxyl Groups of Sub... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2d09 | ||||||
|---|---|---|---|---|---|---|---|
| Title | A Role for Active Site Water Molecules and Hydroxyl Groups of Substrate for Oxygen Activation in Cytochrome P450 158A2 | ||||||
 Components Components | putative cytochrome P450 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / streptomyces / Cytochrome P450 oxidoreductase / CYP158A2 / proton transfer / dioxygen activation | ||||||
| Function / homology |  Function and homology information Function and homology informationbiflaviolin synthase / pigment metabolic process / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen / monooxygenase activity / iron ion binding / heme binding Similarity search - Function | ||||||
| Biological species |  Streptomyces coelicolor (bacteria) Streptomyces coelicolor (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / structure refinement with starting model / Resolution: 1.8 Å SYNCHROTRON / structure refinement with starting model / Resolution: 1.8 Å | ||||||
 Authors Authors | Zhao, B. / Waterman, M.R. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2005 Journal: J.Biol.Chem. / Year: 2005Title: Role of active site water molecules and substrate hydroxyl groups in oxygen activation by cytochrome P450 158A2: a new mechanism of proton transfer Authors: Zhao, B. / Guengerich, F.P. / Voehler, M. / Waterman, M.R. #1:  Journal: J.Biol.Chem. / Year: 2005 Journal: J.Biol.Chem. / Year: 2005Title: Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2 Authors: Zhao, B. / Guengerich, F.P. / Bellamine, A. / Lamb, D.C. / Izumikawa, M. / Lei, L. / Podust, L.M. / Sundaramoorthy, M. / Kalaitzis, J.A. / Reddy, L.M. / Kelly, S.L. / Moore, B.S. / Stec, D. ...Authors: Zhao, B. / Guengerich, F.P. / Bellamine, A. / Lamb, D.C. / Izumikawa, M. / Lei, L. / Podust, L.M. / Sundaramoorthy, M. / Kalaitzis, J.A. / Reddy, L.M. / Kelly, S.L. / Moore, B.S. / Stec, D. / Voehler, M. / Falck, J.R. / Shimada, T. / Waterman, M.R. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2d09.cif.gz 2d09.cif.gz | 99.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2d09.ent.gz pdb2d09.ent.gz | 74.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2d09.json.gz 2d09.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d0/2d09 https://data.pdbj.org/pub/pdb/validation_reports/d0/2d09 ftp://data.pdbj.org/pub/pdb/validation_reports/d0/2d09 ftp://data.pdbj.org/pub/pdb/validation_reports/d0/2d09 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2d0eC  1t93S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 44621.727 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptomyces coelicolor (bacteria) / Strain: A3(2) / Plasmid: pET17b / Production host: Streptomyces coelicolor (bacteria) / Strain: A3(2) / Plasmid: pET17b / Production host:  References: UniProt: Q9FCA6, Oxidoreductases; Acting on paired donors, with incorporation or reduction of molecular oxygen | ||||
|---|---|---|---|---|---|
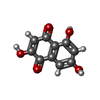
| #2: Chemical | ChemComp-HEM / | ||||
| #3: Chemical | | #4: Chemical | ChemComp-OXY / | #5: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.449 Å3/Da / Density % sol: 47.84 % |
|---|---|
| Crystal grow | Temperature: 295 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: PEG 3350, flaviolin, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 295K |
-Data collection
| Diffraction | Mean temperature: 200 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 22-BM / Wavelength: 0.95 Å / Beamline: 22-BM / Wavelength: 0.95 Å |
| Detector | Type: BRUKER PROTEUM 225 / Detector: CCD / Date: Jun 23, 2005 |
| Radiation | Monochromator: Si 220 CHANNEL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.95 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→37.4 Å / Num. all: 37024 / Num. obs: 35918 / % possible obs: 97 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Rmerge(I) obs: 0.085 / Rsym value: 0.104 |
| Reflection shell | Resolution: 1.8→1.86 Å / Rmerge(I) obs: 0.352 / Mean I/σ(I) obs: 2.6 / Rsym value: 0.336 / % possible all: 89.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: structure refinement with starting model Starting model: PDB ENTRY 1T93 Resolution: 1.8→15 Å / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||
| Solvent computation | Bsol: 49.895 Å2 | ||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.811 Å2
| ||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→15 Å
| ||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.88 Å / Rfactor Rfree error: 0.012
| ||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj