[English] 日本語
 Yorodumi
Yorodumi- PDB-2cjt: Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Hetero... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2cjt | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | ||||||
 Components Components | UNC-13 HOMOLOG A | ||||||
 Keywords Keywords | EXOCYTOSIS / PHORBOL-ESTER BINDING / NEUROTRANSMITTER RELEASE / RIM / MUNC13 / C2 DOMAINS / METAL-BINDING / PROTEIN-PROTEIN INTERACTIONS / ZINC FINGER / SYNAPTOSOME | ||||||
| Function / homology |  Function and homology information Function and homology informationdense core granule priming / neuronal dense core vesicle exocytosis / diacylglycerol binding / synaptic vesicle maturation / synaptic vesicle docking / regulation of synaptic vesicle priming / positive regulation of synaptic plasticity / positive regulation of dendrite extension / positive regulation of glutamate receptor signaling pathway / regulation of short-term neuronal synaptic plasticity ...dense core granule priming / neuronal dense core vesicle exocytosis / diacylglycerol binding / synaptic vesicle maturation / synaptic vesicle docking / regulation of synaptic vesicle priming / positive regulation of synaptic plasticity / positive regulation of dendrite extension / positive regulation of glutamate receptor signaling pathway / regulation of short-term neuronal synaptic plasticity / neurotransmitter secretion / innervation / regulation of amyloid precursor protein catabolic process / syntaxin binding / presynaptic active zone / syntaxin-1 binding / Golgi-associated vesicle / spectrin binding / positive regulation of neurotransmitter secretion / neuromuscular junction development / synaptic vesicle priming / excitatory synapse / synaptic vesicle exocytosis / amyloid-beta metabolic process / calyx of Held / synaptic membrane / SNARE binding / synaptic transmission, glutamatergic / neuromuscular junction / phospholipid binding / long-term synaptic potentiation / terminal bouton / synaptic vesicle membrane / presynapse / presynaptic membrane / cell differentiation / calmodulin binding / protein domain specific binding / axon / calcium ion binding / synapse / protein-containing complex binding / glutamatergic synapse / protein-containing complex / zinc ion binding / identical protein binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.44 Å MOLECULAR REPLACEMENT / Resolution: 1.44 Å | ||||||
 Authors Authors | Lu, J. / Machius, M. / Dulubova, I. / Dai, H. / Sudhof, T.C. / Tomchick, D.R. / Rizo, J. | ||||||
 Citation Citation |  Journal: Plos Biol. / Year: 2006 Journal: Plos Biol. / Year: 2006Title: Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch Authors: Lu, J. / Machius, M. / Dulubova, I. / Dai, H. / Sudhof, T.C. / Tomchick, D.R. / Rizo, J. #1:  Journal: Embo J. / Year: 2005 Journal: Embo J. / Year: 2005Title: A Munc13-Rim-Rab3 Tripartite Complex: From Priming to Plasticity Authors: Dulubova, I. / Lou, X. / Lu, J. / Huryeva, I. / Alam, A. / Schneggenburger, R. / Sudhof, T.T. / Rizo, J. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2cjt.cif.gz 2cjt.cif.gz | 132.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2cjt.ent.gz pdb2cjt.ent.gz | 105 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2cjt.json.gz 2cjt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2cjt_validation.pdf.gz 2cjt_validation.pdf.gz | 480.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2cjt_full_validation.pdf.gz 2cjt_full_validation.pdf.gz | 485.4 KB | Display | |
| Data in XML |  2cjt_validation.xml.gz 2cjt_validation.xml.gz | 29.1 KB | Display | |
| Data in CIF |  2cjt_validation.cif.gz 2cjt_validation.cif.gz | 43.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cj/2cjt https://data.pdbj.org/pub/pdb/validation_reports/cj/2cjt ftp://data.pdbj.org/pub/pdb/validation_reports/cj/2cjt ftp://data.pdbj.org/pub/pdb/validation_reports/cj/2cjt | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 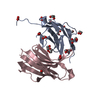
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14761.839 Da / Num. of mol.: 4 / Fragment: C2A DOMAIN, RESIDUES 1-128 Source method: isolated from a genetically manipulated source Details: THE RECOMBINANT PROTEIN CONTAINS RESIDUES 1-128 OF MUNC13-1 AND VECTOR-DERIVED SEQUENCES, GGV- AT THE N-TERMINUS Source: (gene. exp.)   #2: Chemical | ChemComp-EDO / #3: Chemical | ChemComp-FMT / #4: Water | ChemComp-HOH / | Sequence details | THE RECOMBINANT PROTEIN CONTAINS RESIDUES 1-128 OF MUNC13- 1 AND VECTOR-DERIVED SEQUENCES, GGV- AT ...THE RECOMBINAN | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 48.3 % Description: FOR MOLECULAR REPLACEMENT, INITIAL MODEL COORDINATES WERE OBTAINED BY MODIFYING THE COORDINATES OF THE RAT MUNC13-1 C2B-DOMAIN DERIVED FROM OUR UNPUBLISHED RESULTS. |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 4.5 Details: VAPOR DIFFUSION; HANGING DROP; PROTEIN: 12 MG/ML MUNC13-1 IN 30 MM TRIS, 150 MM NACL AND 1 MM TCEP, PH 7.4; RESERVOIR: 0.4 M MAGNESIUM FORMATE, 0.1 M SODIUM ACETATE (PH 4.5); DROP: 1 ...Details: VAPOR DIFFUSION; HANGING DROP; PROTEIN: 12 MG/ML MUNC13-1 IN 30 MM TRIS, 150 MM NACL AND 1 MM TCEP, PH 7.4; RESERVOIR: 0.4 M MAGNESIUM FORMATE, 0.1 M SODIUM ACETATE (PH 4.5); DROP: 1 MICROLITER PROTEIN PLUS 1 MICROLITER RESERVOIR; TEMPERATURE: 20 DEGREES CELSIUS; CRYSTALS APPEARED OVERNIGHT AND GREW TO A FINAL SIZE OF ABOUT 0.05 MM X 0.05 MM X 0.35 MM WITHIN 3 DAYS. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-BM / Wavelength: 0.97959 / Beamline: 19-BM / Wavelength: 0.97959 |
| Detector | Type: CUSTOM / Detector: CCD / Date: Oct 20, 2004 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97959 Å / Relative weight: 1 |
| Reflection | Resolution: 1.44→47.12 Å / Num. obs: 99080 / % possible obs: 99.7 % / Observed criterion σ(I): -3 / Redundancy: 4.3 % / Biso Wilson estimate: 14.4 Å2 / Rmerge(I) obs: 0.06 / Net I/σ(I): 24.1 |
| Reflection shell | Resolution: 1.44→1.47 Å / Redundancy: 2.9 % / Rmerge(I) obs: 0.36 / Mean I/σ(I) obs: 3 / % possible all: 97.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT / Resolution: 1.44→20 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.958 / SU B: 1.984 / SU ML: 0.039 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.058 / ESU R Free: 0.061 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. MOLECULAR REPLACEMENT / Resolution: 1.44→20 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.958 / SU B: 1.984 / SU ML: 0.039 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.058 / ESU R Free: 0.061 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.17 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.44→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj









