+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1s9l | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | NMR Solution Structure of a Parallel LNA Quadruplex | ||||||||||||||||||
 Components Components | 5'-((TLN)P* Keywords KeywordsRNA / LNA / QUADRUPLEX | Function / homology | RNA |  Function and homology information Function and homology informationMethod | SOLUTION NMR / torsion angle dynamics |  Authors AuthorsRandazzo, A. / Esposito, V. / Ohlenschlager, O. / Ramachandran, R. / Mayola, L. |  Citation Citation Journal: Nucleic Acids Res. / Year: 2004 Journal: Nucleic Acids Res. / Year: 2004Title: NMR solution structure of a parallel LNA quadruplex. Authors: Randazzo, A. / Esposito, V. / Ohlenschlager, O. / Ramachandran, R. / Mayola, L. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1s9l.cif.gz 1s9l.cif.gz | 263.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1s9l.ent.gz pdb1s9l.ent.gz | 228.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1s9l.json.gz 1s9l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s9/1s9l https://data.pdbj.org/pub/pdb/validation_reports/s9/1s9l ftp://data.pdbj.org/pub/pdb/validation_reports/s9/1s9l ftp://data.pdbj.org/pub/pdb/validation_reports/s9/1s9l | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 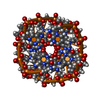
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 1691.098 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: This sequence contains all LNA residues |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: This structure was determined using standard 2D homonuclear techniques |
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 7 / Pressure: ambient / Temperature: 300 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | ||||||||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 Details: 292 (73 per strand) NOE-derived distance constraints, 48 hydrogen bonds constraints, 36 (9 per strand) dihedral angle restraints. The 20 structures with the lowest CYANA target functions ...Details: 292 (73 per strand) NOE-derived distance constraints, 48 hydrogen bonds constraints, 36 (9 per strand) dihedral angle restraints. The 20 structures with the lowest CYANA target functions were subjected energy minimization (with no angle constraints) using the conjugate gradient method and the CVFF force field | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 20 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller







 PDBj
PDBj CYANA
CYANA