[English] 日本語
 Yorodumi
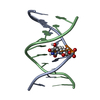
Yorodumi- PDB-1s75: SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC A... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1s75 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / DNA DOUBLE HELIX WITH ENLARGED MINER GROOVE AND HELICAL KINK | ||||||
| Function / homology | DNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / TORSION ANGLE DYNAMICS, MOLECULAR DYNAMICS | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Aramini, J.M. / Cleaver, S.H. / Pon, R.T. / Cunningham, R.P. / Germann, M.W. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2004 Journal: J.Mol.Biol. / Year: 2004Title: Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV. Authors: Aramini, J.M. / Cleaver, S.H. / Pon, R.T. / Cunningham, R.P. / Germann, M.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1s75.cif.gz 1s75.cif.gz | 22.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1s75.ent.gz pdb1s75.ent.gz | 14.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1s75.json.gz 1s75.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s7/1s75 https://data.pdbj.org/pub/pdb/validation_reports/s7/1s75 ftp://data.pdbj.org/pub/pdb/validation_reports/s7/1s75 ftp://data.pdbj.org/pub/pdb/validation_reports/s7/1s75 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 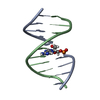
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 3029.994 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: THE CORE OF THE SEQUENCE CORRESPONDS TO THE RECOGNITION SITE OF E. COLI ENDONUCLEASE IV |
|---|---|
| #2: DNA chain | Mass: 3061.003 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||||||
| NMR details | Text: FOR NON-EXCHANGEABLE PROTONS, NOESY EXPERIMENTS IN D2O WERE PERFORMED WITH 10S RELAXATION DELAYS AND MIXING TIMES OF 75MS, 150MS AND 250 MS. FOR EXCHANGEABLE PROTONS, A WATERGATE NOESY WAS ...Text: FOR NON-EXCHANGEABLE PROTONS, NOESY EXPERIMENTS IN D2O WERE PERFORMED WITH 10S RELAXATION DELAYS AND MIXING TIMES OF 75MS, 150MS AND 250 MS. FOR EXCHANGEABLE PROTONS, A WATERGATE NOESY WAS PERFORMED WITH A RELAXATION DELAY OF 4S AND 150MS MIXING TIME. |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: TORSION ANGLE DYNAMICS, MOLECULAR DYNAMICS / Software ordinal: 1 Details: THE STRUCTURE IS BASED ON A TOTAL OF 502 RESTRAINTS: 284 DISTANCE RESTRAINTS, 80 ENDOCYCLIC TORSION ANGLE RESTRAINTS, 50 WATSON-CRICK DISTANCE AND ANGLE RESTRAINTS, AND 88 BACKBONE TORSION ...Details: THE STRUCTURE IS BASED ON A TOTAL OF 502 RESTRAINTS: 284 DISTANCE RESTRAINTS, 80 ENDOCYCLIC TORSION ANGLE RESTRAINTS, 50 WATSON-CRICK DISTANCE AND ANGLE RESTRAINTS, AND 88 BACKBONE TORSION ANGLE RESTRAINTS. THE ALPHAA DUPLEX STRUCTURE WAS ELUCIDATED BY A COMBINATION OF DYANA AND RMD/REM IN AMBER. ALL CALCULATIONS WERE PERFORMED IN VACUO. THE FINAL STRUCTURE DEPOSITED HERE WAS OBTAINED BY COORDINATE AVERAGING THE FINAL ENSEMBLE OF 10 RMD/REM STRUCTURES FOLLOWED BY RESTRAINED ENERGY MINIMIZATION. THE IN VACUO STRUCTURE WAS THEN SOLVATED IN A SOLVENT BOX AND NA+ COUNTERIONS. THE FOLLOWING SIMULATIONS CONTAINED 25 C-H RESIDUAL DIPOLAR COUPLINGS RESTRAINTS. THE FINAL SOLVATED STRUCTURE WAS OBTAINED BY AVERAGING THE LAST 5PS OF A 1 NS RMD RUN FOLLOWED BY REM. | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller




 PDBj
PDBj

 HSQC
HSQC