+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1qyp | ||||||
|---|---|---|---|---|---|---|---|
| Title | THERMOCOCCUS CELER RPB9, NMR, 25 STRUCTURES | ||||||
 Components Components | RNA POLYMERASE II | ||||||
 Keywords Keywords | TRANSCRIPTION / RNA POLYMERASE II SUBUNIT / RPB9 / ZN RIBBON / HYPERTHERMOPHILIC / EXTREMOPHILE | ||||||
| Function / homology |  Function and homology information Function and homology informationDNA-directed RNA polymerase complex / DNA-directed RNA polymerase activity / DNA-templated transcription / regulation of DNA-templated transcription / DNA binding / zinc ion binding Similarity search - Function | ||||||
| Biological species |   Thermococcus celer (archaea) Thermococcus celer (archaea) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Wang, B. / Jones, D.N.M. / Kaine, B.P. / Weiss, M.A. | ||||||
 Citation Citation |  Journal: Structure / Year: 1998 Journal: Structure / Year: 1998Title: High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Authors: Wang, B. / Jones, D.N. / Kaine, B.P. / Weiss, M.A. #1:  Journal: Nature / Year: 1995 Journal: Nature / Year: 1995Title: Erratum. Structure of a New Nucleic-Acid-Binding Motif in Eukaryotic Transcriptional Elongation Factor Tfiis Authors: Qian, X. / Jeon, C. / Yoon, H. / Agarwal, K. / Weiss, M.A. #2:  Journal: Nature / Year: 1993 Journal: Nature / Year: 1993Title: Structure of a New Nucleic-Acid-Binding Motif in Eukaryotic Transcriptional Elongation Factor Tfiis Authors: Qian, X. / Jeon, C. / Yoon, H. / Agarwal, K. / Weiss, M.A. | ||||||
| History |
|
- Structure visualization
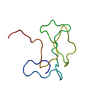
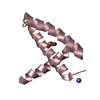
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1qyp.cif.gz 1qyp.cif.gz | 442.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1qyp.ent.gz pdb1qyp.ent.gz | 369 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1qyp.json.gz 1qyp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qy/1qyp https://data.pdbj.org/pub/pdb/validation_reports/qy/1qyp ftp://data.pdbj.org/pub/pdb/validation_reports/qy/1qyp ftp://data.pdbj.org/pub/pdb/validation_reports/qy/1qyp | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6642.467 Da / Num. of mol.: 1 / Fragment: SUBUNIT RPB9, C-TERMINAL DOMAIN, RESIDUES 58 -110 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Thermococcus celer (archaea) / Cell line: BL21 / Plasmid: PET15-B / Production host: Thermococcus celer (archaea) / Cell line: BL21 / Plasmid: PET15-B / Production host:  |
|---|---|
| #2: Chemical | ChemComp-ZN / |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Sample conditions | pH: 6 / Temperature: 313 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||||||
| NMR ensemble | Conformers submitted total number: 25 |
 Movie
Movie Controller
Controller









 PDBj
PDBj




