[English] 日本語
 Yorodumi
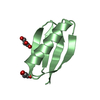
Yorodumi- PDB-1pgb: TWO CRYSTAL STRUCTURES OF THE B1 IMMUNOGLOBULIN-BINDING DOMAIN OF... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pgb | ||||||
|---|---|---|---|---|---|---|---|
| Title | TWO CRYSTAL STRUCTURES OF THE B1 IMMUNOGLOBULIN-BINDING DOMAIN OF STREPTOCCOCAL PROTEIN G AND COMPARISON WITH NMR | ||||||
 Components Components | PROTEIN G | ||||||
 Keywords Keywords | IMMUNOGLOBULIN BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Streptococcus sp. GX7805 (bacteria) Streptococcus sp. GX7805 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.92 Å X-RAY DIFFRACTION / Resolution: 1.92 Å | ||||||
 Authors Authors | Gallagher, T. / Alexander, P. / Bryan, P. / Gilliland, G.L. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Authors: Gallagher, T. / Alexander, P. / Bryan, P. / Gilliland, G.L. #1:  Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: 1.67 Angstroms X-Ray Structure of the B2 Immunoglobulin-Binding Domain of Streptoccocal Protein G and Comparison to the NMR Structure of the B1 Domain Authors: Achari, A. / Hale, S.P. / Howard, A.J. / Clore, G.M. / Gronenborn, A.M. / Hardman, K.D. / Whitlow, M. #2:  Journal: Science / Year: 1991 Journal: Science / Year: 1991Title: A Novel, Highly Stable Fold of the Immunoglobulin Binding Domain of Streptococcal Protein G Authors: Gronenborn, A.M. / Filpula, D.R. / Essig, N.Z. / Achari, A. / Whitlow, M. / Wingfield, P.T. / Clore, G.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pgb.cif.gz 1pgb.cif.gz | 20.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pgb.ent.gz pdb1pgb.ent.gz | 12.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pgb.json.gz 1pgb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pg/1pgb https://data.pdbj.org/pub/pdb/validation_reports/pg/1pgb ftp://data.pdbj.org/pub/pdb/validation_reports/pg/1pgb ftp://data.pdbj.org/pub/pdb/validation_reports/pg/1pgb | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6201.784 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Streptococcus sp. GX7805 (bacteria) / References: UniProt: P06654 Streptococcus sp. GX7805 (bacteria) / References: UniProt: P06654 |
|---|---|
| #2: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.48 Å3/Da / Density % sol: 50.31 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 18 ℃ / pH: 4 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.86 Å / Num. obs: 5014 / Rmerge(I) obs: 0.068 |
| Reflection shell | *PLUS Mean I/σ(I) obs: 4.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.92→6 Å / σ(I): 1 /
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.92→6 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.198 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller








 PDBj
PDBj
