[English] 日本語
 Yorodumi
Yorodumi- PDB-1par: DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1par | ||||||
|---|---|---|---|---|---|---|---|
| Title | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION/DNA / PROTEIN-DNA COMPLEX / DOUBLE HELIX / TRANSCRIPTION-DNA COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Enterobacteria phage P22 (virus) Enterobacteria phage P22 (virus) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.6 Å X-RAY DIFFRACTION / Resolution: 2.6 Å | ||||||
 Authors Authors | Raumann, B.E. / Rould, M.A. / Pabo, C.O. / Sauer, R.T. | ||||||
 Citation Citation |  Journal: Nature / Year: 1994 Journal: Nature / Year: 1994Title: DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Authors: Raumann, B.E. / Rould, M.A. / Pabo, C.O. / Sauer, R.T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1par.cif.gz 1par.cif.gz | 78.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1par.ent.gz pdb1par.ent.gz | 57.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1par.json.gz 1par.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pa/1par https://data.pdbj.org/pub/pdb/validation_reports/pa/1par ftp://data.pdbj.org/pub/pdb/validation_reports/pa/1par ftp://data.pdbj.org/pub/pdb/validation_reports/pa/1par | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 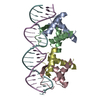
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||
| Unit cell |
| ||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
| ||||||||||||||||
| Details | THE TRANSFORMATION PRESENTED ON *MTRIX 1* RECORDS BELOW WILL YIELD APPROXIMATE COORDINATES FOR CHAIN A WHEN APPLIED TO CHAIN B (DIMER SYMMETRY AXIS). THE TRANSFORMATION PRESENTED ON *MTRIX 2* RECORDS BELOW WILL YIELD APPROXIMATE COORDINATES FOR CHAIN C WHEN APPLIED TO CHAIN D (DIMER SYMMETRY AXIS). THE TRANSFORMATION PRESENTED ON *MTRIX 3* RECORDS BELOW WILL YIELD APPROXIMATE COORDINATES FOR CHAINS A AND B WHEN APPLIED TO CHAINS C AND D, RESPECTIVELY. THIS THIRD TRANSFORMATION RELATES DIMER CD AND THE RIGHT OPERATOR HALF-SITE TO DIMER AB AND THE LEFT OPERATOR HALF-SITE. (TETRAMER SYMMETRY AXIS). |
- Components
Components
| #1: DNA chain | Mass: 6756.387 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
|---|---|---|---|
| #2: DNA chain | Mass: 6743.404 Da / Num. of mol.: 1 / Source method: obtained synthetically | ||
| #3: Protein | Mass: 6238.272 Da / Num. of mol.: 4 / Source method: isolated from a natural source / Source: (natural)  Enterobacteria phage P22 (virus) / Genus: P22-like viruses / References: UniProt: P03050 Enterobacteria phage P22 (virus) / Genus: P22-like viruses / References: UniProt: P03050#4: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.49 Å3/Da / Density % sol: 50.67 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 7.4 / Details: pH 7.40, VAPOR DIFFUSION, HANGING DROP | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 7.4 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE |
|---|---|
| Radiation | Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
| Reflection | Highest resolution: 2.6 Å / Num. all: 161619 / Num. obs: 11253 |
| Reflection | *PLUS Highest resolution: 2.6 Å / % possible obs: 95 % / Num. measured all: 161619 / Rmerge(I) obs: 0.114 |
- Processing
Processing
| Software | Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.6→21 Å / σ(F): 0 /
| ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→21 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.6 Å / Lowest resolution: 21 Å / Rfactor obs: 0.225 / Rfactor Rwork: 0.225 | ||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller












 PDBj
PDBj







































